Difference between revisions of "Ajvree Week 13"
(→11/21/13: import times, results) |
(→11/21/13: table analysis) |
||
| (2 intermediate revisions by one user not shown) | |||
| Line 81: | Line 81: | ||
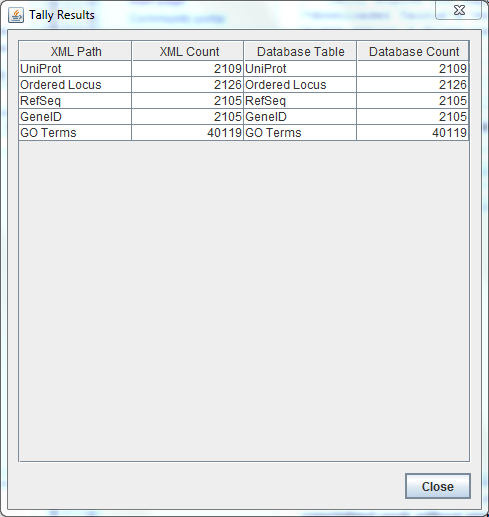
**GeneId's now visible, total of 2105 in both xml and database counts | **GeneId's now visible, total of 2105 in both xml and database counts | ||
**orderedlocusnames still at same value of 2126 in both xml and database counts | **orderedlocusnames still at same value of 2126 in both xml and database counts | ||
| + | **screenshot: [[Image:20131121 TallyEngineE3 tATK TIGR4 AJV.PNG]] | ||
| + | |||
| + | |||
| + | '''Original Row Counts for Export 3''' | ||
| + | *redid row counts using the export 3 file | ||
| + | *compared with benchmark file | ||
| + | *both files had identical number of tables with same categories in each, although some were out of order. | ||
| + | *Screenshots: | ||
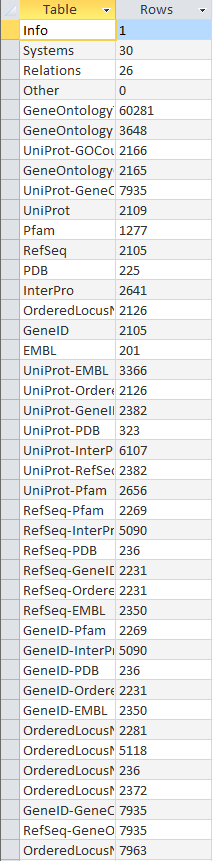
| + | E3 TIGR4: [[Image:20131121 E3rowcounts tATK TIGR4 AJV.PNG]] | ||
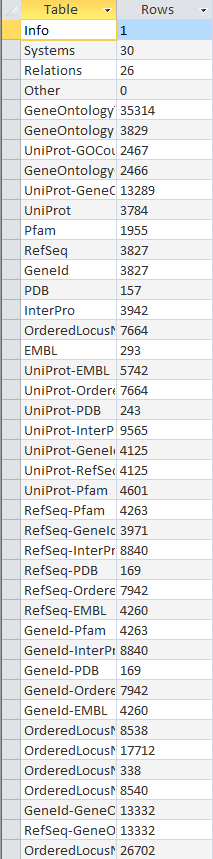
| + | Benchmark:[[Image:20131121 benchmarkrowcounts tATK TIGR4 AJV.PNG]] | ||
| + | |||
| + | '''Table Analysis''' | ||
| + | *looked at tables within E3 gdb file to find inconsistencies in data | ||
| + | |||
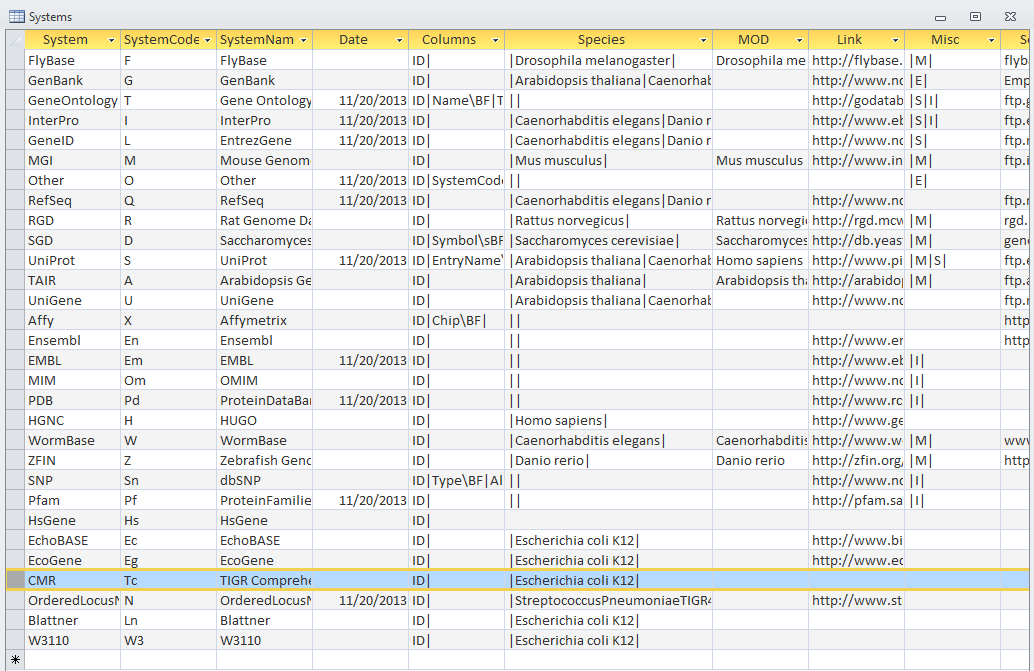
| + | '''Systems Table'''<br> | ||
| + | [[Image:20131121 E3Systemstable tATK TIGR4 AJV.PNG]] | ||
| + | *There are missing dates for quite a few gene ID systems | ||
| + | |||
| + | '''OrderedLocusNames Table''' | ||
| + | *All ID's took the expected form, SP_#### | ||
| + | |||
| + | '''UniProt Table''' | ||
| + | *ID's are scattered. Have a general pattern of beginning with P or Q, following with five characters (mix of numbers and letters) | ||
| + | |||
| + | '''RefSeq Table''' | ||
| + | *all ID's in form NP_###### | ||
==Links== | ==Links== | ||
{{Team ATK}} | {{Team ATK}} | ||
Latest revision as of 18:56, 21 November 2013
Contents |
[edit] Week 12 Information
Export Counting: -open Access, open file (remember to change all to all files) TIGR4 file:
ids used: SP_####
orderedlocusnames count total: 2126 entries
R6 file:
orderedlocusnames count total: 2115 entries
ids used: SPG_####
G54 file:
ids used: SPG_####
orderedlocusnames count total: 2115 entries
After finding identical results for R6 and G54 files, realized that R6 file was actually for the G54 strain. Checked a few times (reopened files multiple times to confirm)
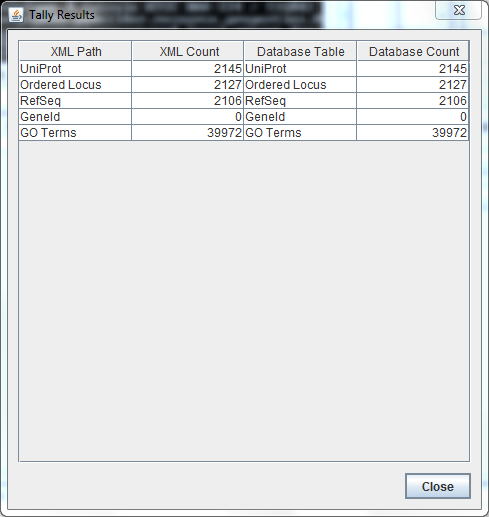
First try on Tally Engine for TIGR4:
XML Count:
orderedlocus: 2127
refseq: 2106
Database Count:
ordered locus: 3831
refseq: 3403
[edit] Week 13
Tally Engine:
- created new database in pgadmin III
- in sql, opened gmbuilder.sql
- ran query, database tables were inserted
- went in to tally engine and imported files
- Xml import took 5.41 min
- GOA import took 0.07 min
- unzipped go-xml file
- OBO-XML import time: 19.92 min
- additional gene ontology information was processed, this took 14.96 min
- ran tally, came up with error
- refreshed gmbuilder and tried again successfully
XMLpipedb Match
- downloaded program from sourceforge
- opened cmd program
- cd Downloads file
- moved xmlmatch jar file to download folder
- used match to look for pattern SP_[0-9][0-9][0-9][0-9]
- Total unique matches: 2126
- almost identical to tally engine results of 2127, minus one result
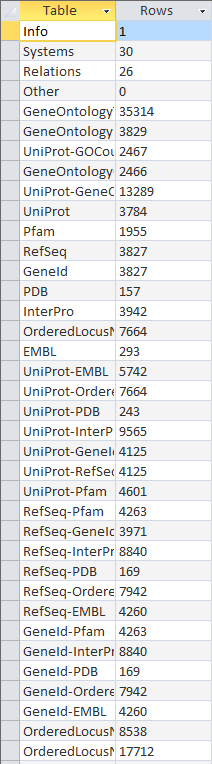
OriginalRowCounts
- Looked at TIGR4 gdb file and benchmard VD file for table similarities/differences
- seemed to have same tables/same information
- took screenshots of both, included here:
- Note: a few of the rows are missing in the benchmark screenshot- could not fit all of them on screen.
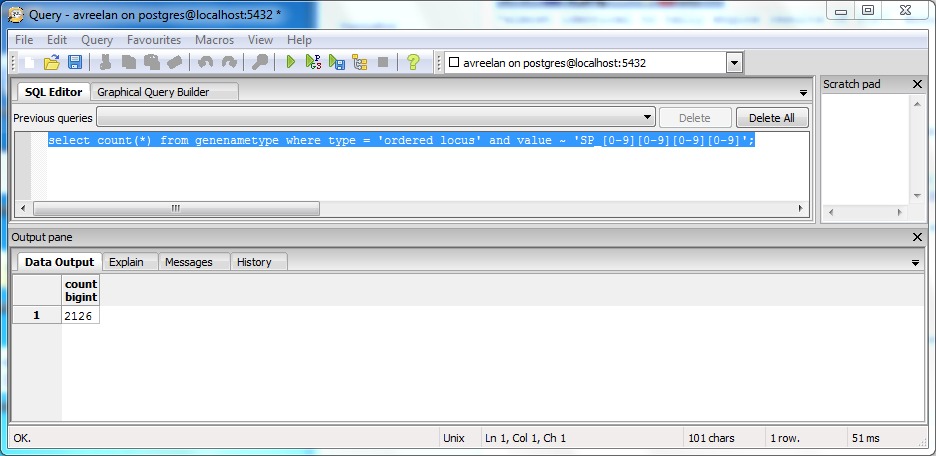
SQL
- used following query to search for matches:
- select count(*) from genenametype where type = 'ordered locus' and value ~ 'SP_[0-9][0-9][0-9][0-9]';
- Result given was 2126
[edit] 11/21/13
Tally Engine for Export 3
- downloaded Taurus's version of gmbuilder to redo tally engine counting
- used export 3 files instead of previous export 1 files
- connected to avreelan database in pgadminIII, inserted new gmbuilder tables
- opened new version of gmbuilder/tally engine, connected to avreelan database
- XML file import took: 2.02 min
- OBO-XML file import took: 6.25 min
- additional gene ontology data processing took: 4.81 min
- GOA file import took: 0.04 min
- Results:
Original Row Counts for Export 3
- redid row counts using the export 3 file
- compared with benchmark file
- both files had identical number of tables with same categories in each, although some were out of order.
- Screenshots:
Table Analysis
- looked at tables within E3 gdb file to find inconsistencies in data
- There are missing dates for quite a few gene ID systems
OrderedLocusNames Table
- All ID's took the expected form, SP_####
UniProt Table
- ID's are scattered. Have a general pattern of beginning with P or Q, following with five characters (mix of numbers and letters)
RefSeq Table
- all ID's in form NP_######
[edit] Links
| Alina's User Page | Kevin's User Page | Tauras's User Page |
| Biological Databases Class Page | Gene Database Project | Gene Database Project Report Guidelines |
- Import Export Cycle 1: tATK Export One: TIGR4 Testing Report
- Import Export Cycle 2: tATK E2: TIGR4 Testing Report
- Import Export Cycle 3: tATK E3: TIGR4 Testing Report
- Import Export Cycle 4: tATK E4: TIGR4 Testing Report
| Project Roles: | Project Manager | Coder | GenMAPP User | Quality Assurance |