Difference between revisions of "TATK E4: TIGR4 Testing Report"
(→Visual Inspection: update refseq info and image) |
(→Compare Gene Database to Outside Resource) |
||
| (9 intermediate revisions by 2 users not shown) | |||
| Line 26: | Line 26: | ||
**Finished by 23:50 | **Finished by 23:50 | ||
* Upload your file and link to it here. [[Media:Streptococcus_pneumoniae_TIGR4_20131125.gdb|Streptococcus_pneumoniae_TIGR4_20131125.gdb]] | * Upload your file and link to it here. [[Media:Streptococcus_pneumoniae_TIGR4_20131125.gdb|Streptococcus_pneumoniae_TIGR4_20131125.gdb]] | ||
| − | |||
| − | |||
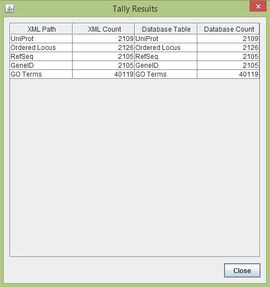
==TallyEngine== | ==TallyEngine== | ||
| Line 34: | Line 32: | ||
**Ordered Locus XML Count: 2126 | **Ordered Locus XML Count: 2126 | ||
**Ordered Locus Database Count: 2126 | **Ordered Locus Database Count: 2126 | ||
| − | |||
[[Image:TallyEngine Capture TPV.JPG|thumb|left|upright=1.5]] | [[Image:TallyEngine Capture TPV.JPG|thumb|left|upright=1.5]] | ||
<br><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><br> | <br><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><BR><br> | ||
| − | |||
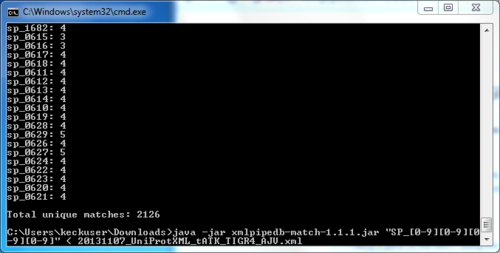
== Using XMLPipeDB match to Validate the XML Results from the TallyEngine== | == Using XMLPipeDB match to Validate the XML Results from the TallyEngine== | ||
| Line 48: | Line 44: | ||
*Total unique matches found: 2126 | *Total unique matches found: 2126 | ||
*This total matched results found in Tally Engine count | *This total matched results found in Tally Engine count | ||
| − | |||
[[Image:20131107 XMLmatch tATK TIGR4 AJV.PNG|500px|]] | [[Image:20131107 XMLmatch tATK TIGR4 AJV.PNG|500px|]] | ||
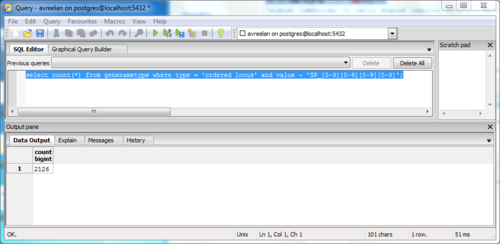
| Line 57: | Line 52: | ||
**''select count(*) from genenametype where type = 'ordered locus' and value ~ 'SP_[0-9][0-9][0-9][0-9]';'' | **''select count(*) from genenametype where type = 'ordered locus' and value ~ 'SP_[0-9][0-9][0-9][0-9]';'' | ||
*Unique matches found: 2126 | *Unique matches found: 2126 | ||
| − | *These results matched those of Tally Engine and XMLPipeDB match, confirming values | + | *These results matched those of Tally Engine and XMLPipeDB match, confirming values<br><br> |
| − | + | ||
[[Image:20131119 SQLcountresults tATK TIGR4 AJV.PNG|500px]] | [[Image:20131119 SQLcountresults tATK TIGR4 AJV.PNG|500px]] | ||
| Line 64: | Line 58: | ||
==OriginalRowCounts Comparison== | ==OriginalRowCounts Comparison== | ||
| − | *Original Row Counts for | + | *Original Row Counts for the gdb file contained had a UniProt Ordered Locus count of 4252 |
| − | * | + | *This was a result of the databases including Gene IDs with and and without underscore |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
==Visual Inspection== | ==Visual Inspection== | ||
| Line 79: | Line 67: | ||
'''OrderedLocusNames Table''' | '''OrderedLocusNames Table''' | ||
| − | * | + | *ID's take the forms SP_#### and SP#### |
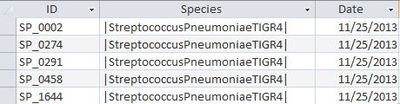
'''UniProt Table''' | '''UniProt Table''' | ||
*ID's are all in expected form SP_#### | *ID's are all in expected form SP_#### | ||
| − | |||
[[Image:Uniprotids2.JPG|400px]] | [[Image:Uniprotids2.JPG|400px]] | ||
| Line 92: | Line 79: | ||
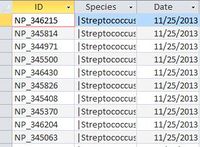
[[Image:Refseqsnip2.JPG|200px]] | [[Image:Refseqsnip2.JPG|200px]] | ||
| − | |||
| − | |||
==.gdb Use in GenMAPP== | ==.gdb Use in GenMAPP== | ||
| Line 131: | Line 116: | ||
[[Image:Ensembl snip.JPG|150px]] | [[Image:Ensembl snip.JPG|150px]] | ||
| − | |||
| − | |||
==Template== | ==Template== | ||
{{Team ATK}} | {{Team ATK}} | ||
Latest revision as of 20:18, 13 December 2013
[edit] Export Information
Version of GenMAPP Builder: 2.0b73
- Database called: tATK_TIGR4_2013NOV25
Computer on which export was run: Tauras' Personal Computer
Postgres Database name: tATK_TIGR4_2013NOV25
UniProt XML filename: 20131118_UniProtXML_tATK_TIGR4_TPV.xml
- UniProt XML version: UniProt Release 2013_11; 2013Nov13
- Time taken to import: 3.15min
GO OBO-XML filename: 20131120_OBOXML_tATK_TPV.obo-xml
- GO OBO-XML version: 2013Nov20
- Time taken to import: 10.59min
- Time taken to process: 9.25min
GOA filename: 20131118_GOA_tATK_TIGR4_TPV.goa
- GOA version: 2013Nov12 14:49
- Time taken to import: 0.03min
Name of .gdb file: Streptococcus_pneumoniae_TIGR4_20131125.gdb
- Time taken to export .gdb: less than 1 hour
- Started at 22:53
- Finished by 23:50
- Upload your file and link to it here. Streptococcus_pneumoniae_TIGR4_20131125.gdb
[edit] TallyEngine
- Tally Engine run on Tauras' personal computer.
- Final Results:
- Ordered Locus XML Count: 2126
- Ordered Locus Database Count: 2126
[edit] Using XMLPipeDB match to Validate the XML Results from the TallyEngine
- XMLPipeDB match program was downloaded from Sourceforge [[1]]
- Moved xmlmatch jar file to Downloads folder on personal computer
- Ran cmd program on personal computer
- Ran query: cd Downloads file
- Searched for pattern: SP_[0-9][0-9][0-9][0-9]
- Total unique matches found: 2126
- This total matched results found in Tally Engine count
[edit] Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine
- Ran pgAdmin III through personal computer to run SQL query
- Command used:
- select count(*) from genenametype where type = 'ordered locus' and value ~ 'SP_[0-9][0-9][0-9][0-9]';
- Unique matches found: 2126
- These results matched those of Tally Engine and XMLPipeDB match, confirming values
Follow the instructions on this page to query the PostgreSQL Database.
[edit] OriginalRowCounts Comparison
- Original Row Counts for the gdb file contained had a UniProt Ordered Locus count of 4252
- This was a result of the databases including Gene IDs with and and without underscore
[edit] Visual Inspection
Systems Table
- There are numerous missing dates for the gene ID systems.
OrderedLocusNames Table
- ID's take the forms SP_#### and SP####
UniProt Table
- ID's are all in expected form SP_####
RefSeq Table
- IDs in form NP_######
- This is expected form for RefSeq, refers to protein accession number.
[edit] .gdb Use in GenMAPP
Note:
[edit] Putting a gene on the MAPP using the GeneFinder window
- Try a sample ID from each of the gene ID systems. Open the Backpage and see if all of the cross-referenced IDs that are supposed to be there are there.
Note:
- no criteria met: Q97RY3 (Q97RY3_STRPN) matched with uniprot page
- not found: Q97NJ5 (Q97NJ5_STRPN) matched with uniprot page
- decreased: Q97SJ6 (CPSC_STRPN) matched with uniprot page
- increased: Q97SB2 (Q97SB2_STRPN) matched with uniprot page
[edit] Creating an Expression Dataset in the Expression Dataset Manager
- How many of the IDs were imported out of the total IDs in the microarray dataset? How many exceptions were there? Look in the EX.txt file and look at the error codes for the records that were not imported into the Expression Dataset. Do these represent IDs that were present in the UniProt XML, but were somehow not imported? or were they not present in the UniProt XML?
Note: 4689 out of 5022 IDs were imported. There were 333 exceptions, all due to not being present in UniProt.
[edit] Coloring a MAPP with expression data
Note: increased was colored red, decreased was colored green, no criteria met was colored grey, and not found was colored white
[edit] Running MAPPFinder
Note: MAPPFinder worked successfully for all of the data used in this project.
[edit] Compare Gene Database to Outside Resource
- Could not find downloadable gene ID list on Ensembl, which was used as MOD for the TIGR4 strain.
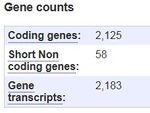
- A 'Coding Gene Count' was given, a value of 2125
- This total is one less than our expected value of 2126
- Inability to find ID list kept us from being able to identify reasons for differences between the values.
[edit] Template
| Alina's User Page | Kevin's User Page | Tauras's User Page |
| Biological Databases Class Page | Gene Database Project | Gene Database Project Report Guidelines |
- Import Export Cycle 1: tATK Export One: TIGR4 Testing Report
- Import Export Cycle 2: tATK E2: TIGR4 Testing Report
- Import Export Cycle 3: tATK E3: TIGR4 Testing Report
- Import Export Cycle 4: tATK E4: TIGR4 Testing Report
| Project Roles: | Project Manager | Coder | GenMAPP User | Quality Assurance |