Difference between revisions of "Vkuehn Week 8"
From LMU BioDB 2013
(added files and images) |
(Reorganized notebook and added details) |
||
| Line 1: | Line 1: | ||
| − | + | =='''Electronic Lab Notebook'''== | |
''''10.10.2013'''' | ''''10.10.2013'''' | ||
#Data accessed from | #Data accessed from | ||
| Line 8: | Line 8: | ||
''''10.1.2013'''' | ''''10.1.2013'''' | ||
Expression dataset manager conveted data resulted in '''[[Media: Merrell_Compiled_Raw_Data_Vibrio_VK_2013.10.15.gex]]''' | Expression dataset manager conveted data resulted in '''[[Media: Merrell_Compiled_Raw_Data_Vibrio_VK_2013.10.15.gex]]''' | ||
| + | ==='''GenMAPP Expression Dataset Procedure'''=== | ||
* Generated lines that resulted in error. During the conversion the data for 2010 resulted in '''121 errors''' | * Generated lines that resulted in error. During the conversion the data for 2010 resulted in '''121 errors''' | ||
* Generated an exception file '''[[Media:Merrell_Compiled_Raw_Data_Vibrio_VK_2013.10.15.EX.txt]]''' | * Generated an exception file '''[[Media:Merrell_Compiled_Raw_Data_Vibrio_VK_2013.10.15.EX.txt]]''' | ||
| Line 18: | Line 19: | ||
*: [Avg_LogFC_all] > 0.25 AND [Pvalue] < 0.05 | *: [Avg_LogFC_all] > 0.25 AND [Pvalue] < 0.05 | ||
*: Uploaded color sets | *: Uploaded color sets | ||
| − | '''MAPPFinder Procedure''' | + | ==='''MAPPFinder Procedure'''=== |
| − | * Launch MAPPFinder and choose color set and criteria, select gene ontology and p value | + | * Launch MAPPFinder and make sure that the database for the correct species is loaded |
| + | *Click "Calculate new results" choose the dataset just created (choose Merrell_Compiled_Raw_Data_Vibrio_VK_2013.10.15.gex) | ||
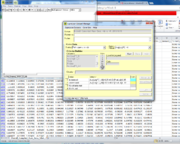
| + | * Choose color set and criteria, select gene ontology and p value (see image to the right) | ||
* Click "run mappfiner" '''[[Media:MerrellVK.mapp]]''' | * Click "run mappfiner" '''[[Media:MerrellVK.mapp]]''' | ||
* Gene Ontology window will open. All of the Gene Ontology terms that have at least 3 genes measured and a p value of less than 0.05 will be highlighted yellow. A term with a p value less than 0.05 is considered a "significant" result. Browse through the tree to see your results. Click on "ranked list" | * Gene Ontology window will open. All of the Gene Ontology terms that have at least 3 genes measured and a p value of less than 0.05 will be highlighted yellow. A term with a p value less than 0.05 is considered a "significant" result. Browse through the tree to see your results. Click on "ranked list" | ||
Revision as of 19:40, 19 October 2013
Electronic Lab Notebook
'10.10.2013'
- Data accessed from
uploaded txt file and excel file:
'10.1.2013' Expression dataset manager conveted data resulted in Media: Merrell_Compiled_Raw_Data_Vibrio_VK_2013.10.15.gex
GenMAPP Expression Dataset Procedure
- Generated lines that resulted in error. During the conversion the data for 2010 resulted in 121 errors
- Generated an exception file Media:Merrell_Compiled_Raw_Data_Vibrio_VK_2013.10.15.EX.txt
- Used autofilter to determine errors. The errors were all gene not found. This was because it was not updated recently so it is ok to proceed. Media:Merrell_Compiled_Raw_Data_Vibrio_VK_2013.10.15.EX.xls
- My partner had a lot more errors than I did. This is because I was using the newer version while he was using the older dataset. Mine had been updated more recently which is why there were less errors because more of the data matched
- Customize the new Expression Dataset by creating new Color Sets which contain the instructions to GenMAPP for displaying data on MAPPs.
- Input the "Avg_LogFC_all" for the Vibrio dataset created in the excel file
- Create criteria builder for "increased":
- [Avg_LogFC_all] > 0.25 AND [Pvalue] < 0.05
- Uploaded color sets
MAPPFinder Procedure
- Launch MAPPFinder and make sure that the database for the correct species is loaded
- Click "Calculate new results" choose the dataset just created (choose Merrell_Compiled_Raw_Data_Vibrio_VK_2013.10.15.gex)
- Choose color set and criteria, select gene ontology and p value (see image to the right)
- Click "run mappfiner" Media:MerrellVK.mapp
- Gene Ontology window will open. All of the Gene Ontology terms that have at least 3 genes measured and a p value of less than 0.05 will be highlighted yellow. A term with a p value less than 0.05 is considered a "significant" result. Browse through the tree to see your results. Click on "ranked list"
- Top 10 Gene Ontology terms (image to the left)
- My terms in comparison with my partners are different, this is because he had different gene dataset that was less updated and there were differences in expression because of this.
- Find the Gene Ontology term(s) with which a particular gene is associated. Search for matched gene from Merrell et al. (2002)VC0647
- List the GO terms associated with each of those genes in your individual journal
- Template:vkuehn
- Viktoria Kuehn
- Week 1 Assignment
- Class Journal 1
- Week 2 Assignment
- Class Journal 2
- Vkuehn Week 2
- Week 3 Assignment
- Class Journal 3
- Vkuehn Week 3
- Week 4 Assignment
- Class Journal 4
- Vkuehn Week 4
- Week 5 Assignment
- Vkuehn Week 5
- Ensembl Database
- Week 6 Assignment
- Class Journal 6
- Vkuehn Week 6
- Week 7 Assignment
- Class Journal 7
- Vkuehn Week 7
- Week 8 Assignment
- Class Journal 8
- Vkuehn Week 8
- Leishmania major
- Week 9 Assignment
- Class Journal 9
- Vkuehn Week 9
- Week 10 Assignment
- Class Journal 10
- Vkuehn Week 10
- Leishmania major
- Week 11 Assignment
- Vkuehn Week 11
- Leishmania major Genome Reference Article Presentation
- Vkuehn Week 12
- Week 12 Assignment
- Leishmania major Week 12 Status Report
- Vkuehn Week 13
- Week 13 Assignment
- Vkuehn Week 15
- Week 15 Assignment
- Vkuehn Individual Assessment and Reflection