Leishmania major
Team Members:
- Kevin McGee : GenMAPP User
- Lena Hunt : Quality Assurance Lena Project Notebook
- Viktoria Kuehn : GenMAPP User
- Gabriel Leis : Coder
Reference Genome Article
- This article was found via the PubMed database and the key terms used were "Leishmania Major [MeSH Terms] AND Genome [Title]"
- This search resulted in 30 articles, about 5 seemed relevant but this one was the first which had the entire genome
- Citation for paper:
- Alasdair C. Ivens, Christopher S. Peacock, Elizabeth A. Worthey, Lee Murphy, Gautam Aggarwa, Matthew Berriman, Ellen Sisk, Marie-Adele Rajandream, Ellen Adlem, Rita Aert, Atashi Anupama, Zina Apostolou, Philip Attipoe, Nathalie Bason, Christopher Bauser, Alfred Beck, Stephen M. Beverley, Gabriella Bianchettin, Katja Borzym, Gordana Bothe, Carlo V. Bruschi, Matt Collins, Eithon Cadag, Laura Ciarloni, Christine Clayton, Richard M. R. Coulson, Ann Cronin, Angela K. Cruz, Robert M. Davies, Javier De Gaudenzi, Deborah E. Dobson, Andreas Duesterhoeft, Gholam Fazelina, Nigel Fosker, Alberto Carlos Frasch, Audrey Fraser, Monika Fuchs, Claudia Gabel, Arlette Goble, André Goffeau, David Harris, Christiane Hertz-Fowler, Helmut Hilbert, David Horn, Yiting Huang, Sven Klages, Andrew Knights, Michael Kube, Natasha Larke, Lyudmila Litvin, Angela Lord, Tin Louie, Marco Marra, David Masuy, Keith Matthews, Shulamit Michaeli, Jeremy C. Mottram, Silke Müller-Auer, Heather Munden, Siri Nelson, Halina Norbertczak, Karen Oliver, Susan O'Neil, Martin Pentony, Thomas M. Poh, Claire Price, Bénédicte Purnelle, Michael A. Quail, Ester Rabbinowitsch, Richard Reinhardt, Michael Rieger, Joel Rinta, Johan Robben, Laura Robertson, Jeronimo C. Ruiz, Simon Rutter, David Saunders, Melanie Schäfer, Jacquie Schein, David C. Schwartz, Kathy Seeger, Amber Seyler, Sarah Sharp, Heesun Shin, Dhileep Sivam, Rob Squares, Steve Squares, Valentina Tosato, Christy Vogt, Guido Volckaert, Rolf Wambutt, Tim Warren, Holger Wedler, John Woodward, Shiguo Zhou, Wolfgang Zimmermann, Deborah F. Smith, Jenefer M. Blackwell, Kenneth D. Stuart, Bart Barrel, Peter J. Myler (2005) The Genome of the Kinetoplastid Parasite, Leishmania major. Science;309(5733):436-42.
- Pdf of the reference article: PDF Reference Article
- The genome sequencing article was used to do a prospective search on the Web of Knowledge database.
- There were 696 results found that referenced this article, the most recent one was in October 2013.
- Many of the most recent articles that cited this paper were involved in studying drug resistance of Leishmania major and studied related pathogenic parasites using sequence comparative analysis, for example.
- The genome sequencing article was used to do a prospective search on the Web of Knowledge database.
- Link to the results page for our reference genome article
- Below is a screen shot of the results.
Leishmania Major Articles
Leishmania Major Data Updates
To update this list edit the following template page: Template:Leishmania Major File Updates
- 11/5/2013
- Media:A-GEOD-6855.adf_A.txt Array Design File
- Media:E-GEOD-10407.idf_A.txt Investigation Description File
- Media:E-GEOD-10407.processed.1_A.zip Processed Data
- Media:E-GEOD-10407.raw.1_A.zip Raw Data
- Media:E-GEOD-10407.sdrf_A.txt Sample and Data Relationship File
- 11/7/2013
- Media:E-GEOD-10407.sdrf_B.txt Leishmania major Chips
- Completed Import/Export Cycle, conducted a Tally Count, opened .gdb file in Microsoft Access and compared original row counts.
- Access was missing 2 genes for GeneID and RefSeq, and we had no GO terms in the Tally or in Access.
- 11/11/2013
- 11/14/13
- Media:E-GEOD-10407.sdrf_C.txt Edited Sample and Data (Ordered based on species and relevant data)
- Media:L.majorCompiledRawData.txt L. major raw compiled data
- Media:L.infantumCompliedRawData(A).txt L. infantum raw compiled data
- 11/19/2013:
- Media:L.majorCompiledRawData_B.txt L. major raw compiled data with adjusted dye swap values
- Media:L.infantumCompliedRawData(B).txt L. infantum raw compiled data with adjusted dye swap values and sample names
- Media:L.majorCompiledRawData_C.txt L. major raw compiled data with adjusted dye swap values and sample names
- 11/21/2013
- 12/3/2013
- File:LeishmaniaCompiledStatAnalysis(A).txt
- File:LeishmaniaCompiledStatAnalysis(A).EX.txt
- Media:L.infantumStats_B.xls Fixed infantum p-value parenthesis
- Media:LeishmaniaCompiledStatAnalysis(B).txt
- Media:LeishmaniaCompiledStatAnalysisLMJFiltered.txt Original File That should work in GenMAPP once coding is fixed
- Media:LeishmaniaCompiledStatAnalysisLMJFiltered(B).txt Quick fix file with underscores to work in GenMAPP until coding is fixed
- Media:LeishmaniaCompiledStatAnalysisLMJFiltered(B).EX.txt
- Media:LeishmaniaCompiledStatAnalysisLMJFiltered(B).gex
- Media:ExceptionFileErrorID.txt
- Leishmania Major Group Project Report
- Media:Dist Leishmania Lena Gabe 05112013.zip
- 12/5/2013
- 12/7/2013
- 12/11/2013
- 12/12/13
- File:LeishmaniaCompiledStatAnalysis(C).txt The data from L. major only for analyzing statistics
Data Links
Leishmania Major Helpful Links
Leishmania Major Home Page
- Leishmania Major Articles
- Template:Leishmania Major Navigation
- Template:Leishmania Major File Updates
- The Plan
Team Members:
- Viktoria (GenMAPP User, Project Manager)
- Gabriel (Coder)
- Lena (Quality Assurance)
- Kevin (GenMAPP User)
Status Reports:
Relevant Project Links:
- Gene Database Project
- Gene Database Project Report Guidelines
- Project Manager
- Coder
- Quality Assurance
- GenMAPP User
Group Projects
Leishmania major Genome Reference Article Presentation
Import/Export
Export Information
- Uniprot: 7.12 minutes
- Media:UniprotXML Leishmania 05112013 Gabe Lena.xml
- GO OBO: 6.32 minutes
- Media:Leishmania 05112013 Gabe Lena.obo-xml.gz
- GOA: 4.54 minutes
- Media:LeishmaniaGOA 05112013 GabeLena.goa
Tally Engine
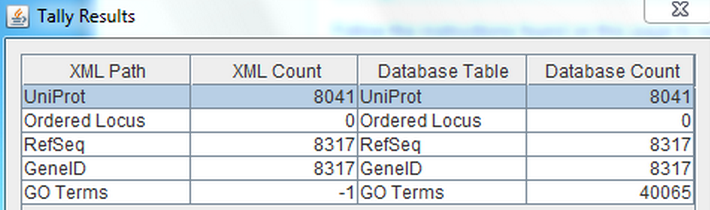
Original Row Counts Comparison
- Uniprot has 8041 which is the same the tallycount.
- There were 0 ordered locus, which is the same as the tallycount.
- There were 8315 hits for RefSeq, which is 2 fewer than was seen in the tallycount.
- There were 8315 hits for GeneID, which is 2 fewer than was seen in the tallycount.
Gene Database Testing Report
Export Information
Version of GenMAPP Builder: gmbuilder2.0-b72
Computer on which export was run: Front row , second computer from the left
Postgres Database name:Leishmania_major_11262013
UniProt XML filename: UniprotXML Leishmania 05112013 Gabe Lena.xml
- UniProt XML version (The version information can be found at the UniProt News Page): UniProt release 2013_10 - October 16, 2013
- Time taken to import: 7.12 minutes
GO OBO-XML filename: Leishmania 05112013 Gabe Lena.obo-xml.gz
- GO OBO-XML version (The version information can be found in the file properties after the file downloaded from the GO Download page has been unzipped): Monday, November 04, 2013, 2:03:38 AM
- Time taken to import:6.32 minutes
- Time taken to process: 0.4 minutes
GOA filename:LeishmaniaGOA 19112013 Lena Gabe.goa
- GOA version (News on this page records past releases; current information can be found in the Last modified field on the FTP site): 14 November, 2013
12-Nov-2013 11:47 3.0M
- Time taken to import: 4.54 minutes
Name of .gdb file: Media:LeishmaniaGDB Lena Gabe 20131203.gdb
- Upload your file and link to it here.
Note:
TallyEngine
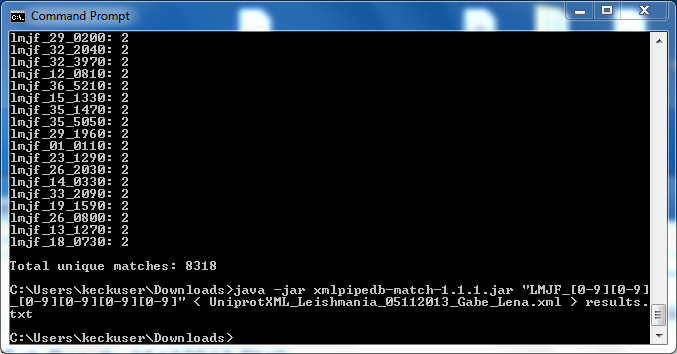
Using XMLPipeDB match to Validate the XML Results from the TallyEngine
Are your results the same as you got for the TallyEngine? Why or why not?
- We found 8353, we are missing two ORFs, but we will try to find them in Postgres Query.
Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine
Follow the instructions on this page to query the PostgreSQL Database.
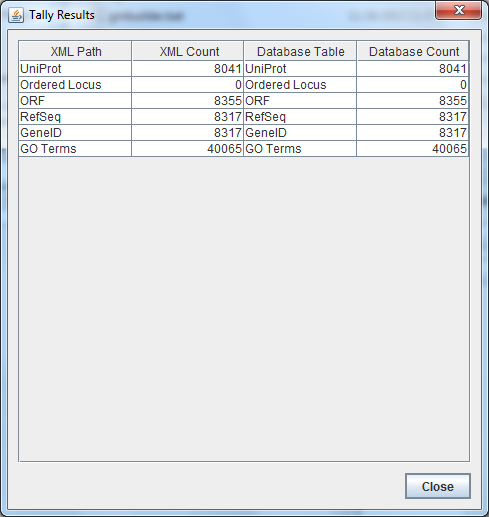
OriginalRowCounts Comparison
Within the .gdb file, look at the OriginalRowCounts table to see if the database has the expected tables with the expected number of records. Compare the tables and records with a benchmark .gdb file.
Benchmark .gdb file: (for the Week 9 Assignment, use the "Vc-Std_External_20101022.gdb" as your benchmark, downloadable from here.
Copy the OriginalRowCounts table and paste it here:
Note:
Visual Inspection
Perform visual inspection of individual tables to see if there are any problems.
- Look at the Systems table. Is there a date in the Date field for all gene ID systems present in the database?
- Open the UniProt, RefSeq, and OrderedLocusNames tables. Scroll down through the table. Do all of the IDs look like they take the correct form for that type of ID?
Note:
.gdb Use in GenMAPP
Note:
Putting a gene on the MAPP using the GeneFinder window
- Try a sample ID from each of the gene ID systems. Open the Backpage and see if all of the cross-referenced IDs that are supposed to be there are there.
Note:
Creating an Expression Dataset in the Expression Dataset Manager
- How many of the IDs were imported out of the total IDs in the microarray dataset? How many exceptions were there? Look in the EX.txt file and look at the error codes for the records that were not imported into the Expression Dataset. Do these represent IDs that were present in the UniProt XML, but were somehow not imported? or were they not present in the UniProt XML?
Note:
Coloring a MAPP with expression data
Note:
Running MAPPFinder
Note: