Bklein7 Week 14
Contents
Overview
GennMAPP Builder version cw20151201: File:Dist cw20151201.zip.
- Build Details: Includes baseline Bordetella pertussis custom class.
- Truncated Gene Database Testing Report: Gene Database Testing Report- cw20151201
GenMAPP Builder version cw20151203: File:Dist cw20151203.zip
- Build Details: Includes Bordetella pertussis custom class expanded to include ORF listings in exports.
- Gene Database Testing Report: Gene Database Testing Report- cw20151203
Bordetella Pertussis Species Profile Creation
To begin the work below, I opened the gmbuilder project within the Java perspective using the program Eclipse. The details regarding the developer rig I used can be found on my Week 12 Assignment Page.
Creating the Species Profile
- I exposed the contents of the src folder.
- I right-clicked on the edu.lmu.xmlpipedb.gmbuilder.databasetoolkit.profiles package and chose New > Class from the popup menu.
- In the dialog that appears, I entered the following:
- Name:
BordetellaPertussisUniProtSpeciesProfile - Superclass:
edu.lmu.xmlpipedb.gmbuilder.databasetoolkit.profiles.UniProtSpeciesProfile
- Name:
- I clicked Finish. This created a new ' file entitled BordetellaPertussisUniProtSpeciesProfile.java within the edu.lmu.xmlpipedb.gmbuilder.databasetoolkit.profiles package.
Customizing the Species Profile
- I opened BordetellaPertussisUniProtSpeciesProfile.java, which appeared in the editor region of Eclipse.
- I overrode the method that supplies the name of the species and the description of the profile by adding the following constructor block right below the public class line in the new file.
public BordetellaPertussisUniProtSpeciesProfile() {
super("Bordetella pertussis", 257313, "This profile customizes the GenMAPP Builder export for " +
"Bordetella pertussis" +
" data loaded from a UniProt XML file.");
}
- To customize the species profile with the species name in the OrderedLocusNames record of the Systems table as well as a link query for that same record, I added the following method block right below the constructor block:
@Override
public TableManager getSystemsTableManagerCustomizations(TableManager tableManager, DatabaseProfile dbProfile) {
super.getSystemsTableManagerCustomizations(tableManager, dbProfile);
tableManager.submit("Systems", QueryType.update, new String[][] {
{ "SystemCode", "N" },
{ "Species", "|" + getSpeciesName() + "|" }
});
tableManager.submit("Systems", QueryType.update, new String[][] {
{ "SystemCode", "N" },
{ "Link", "http://www.genedb.org/gene/~;jsessionid=A06A0EFE93C64E476380393D4CBEFA69?actionName=%2FQuery%2FquickSearch&resultsSize=1&taxonNodeName=Bpertussis" }
});
return tableManager;
}
- At this point, my code had several red error badges. To fix this, I chose Organize Imports from the Source menu.
- I saved the file, at which point the red error badges disappeared.
Adding the Species Profile to the Catalog of Known Species Profiles
In order to have GenMAPP Builder recognize my new species profile, I had to edit an existing file:
- Under edu.lmu.xmlpipedb.gmbuilder.databasetoolkit.profiles, I opened UniProtDatabaseProfile.java.
- Near the top of the file is a block that looks like this:
super("org.uniprot.uniprot.Uniprot",
"This profile defines the requirements "
+ "for any UniProt centric gene database.",
new SpeciesProfile[] {
new EscherichiaColiUniProtSpeciesProfile(),
new ArabidopsisThalianaUniProtSpeciesProfile(),
new PlasmodiumFalciparumUniProtSpeciesProfile(),
new VibrioCholeraeUniprotSpeciesProfile() });
- I added the Bordetella pertussis species profile that I just created to this block. The modified code looked like this:
super("org.uniprot.uniprot.Uniprot",
"This profile defines the requirements "
+ "for any UniProt centric gene database.",
new SpeciesProfile[] {
new EscherichiaColiUniProtSpeciesProfile(),
new ArabidopsisThalianaUniProtSpeciesProfile(),
new PlasmodiumFalciparumUniProtSpeciesProfile(),
new VibrioCholeraeUniprotSpeciesProfile(),
new BordetellaPertussisUniProtSpeciesProfile() });
- I saved my changes. No errors were present in the code.
Creating a New Build of GenMAPP Builder
To create a new version of GenMAPP Builder based on the code I edited in Eclipse, I created a new distribution:
- I opened the gmbuilder project by clicking on the gray triangle to the left of its name.
- I right-clicked on build.xml (within the gmbuilder Java project) and choose Run As > Ant Build... from the menu that appeared.
- In the Edit Configuration dialog that appears, I unchecked dist.
- I checked the clean and dist items in the Targets tab. The Target execution order section near the bottom of the dialog displayed: clean, dist.
- I clicked the Run button.
- When this process completed, I right-clicked on the gmbuilder project folder and chose Refresh from the menu that appeared.
- A dist folder was now visible inside the gmbuilder project folder.
- This corresponded to my personally-built copy of GenMAPP Builder.
To compress the new version of GenMAPP Builder and export it, I navigated to the xmlpipedb folder present on my coding computer. Within this folder, I opened gmbuilder and compressed the dist folder. This compressed file was then uploaded to the wiki here: File:Dist cw20151201.zip.
Testing the New Build of GenMAPP Builder
To test this new build, I downloaded the new distribution (File:Dist cw20151201.zip) on the lab computer and performed the following:
- I ran PostgreSQL (pgAdmin III) and created a new database: bpertussis_cw20151201_gmb3build5.
- I launched the gmbuilder Windows Batch File downloaded from the new dist folder. It opened without complication.
- I began a new import-export cycle using this new version of GenMAPP Builder to see if the Bordetella pertussis custom species I created worked as intended.
- Trial 1- On the final step of the Export Wizard, the Bordetella pertussis species profile available was listed as a generic profile. This demonstrated that the new version of GenMAPP Builder was not coded properly.
- I first troubleshooted by going over my code and verifying that the correct TaxonID for Bordetella pertussis was used. All of the code was indeed correct.
- The issue ended up being that the build I originally exported from my coding computer was outdated and therefore did not include my changes to the gmbuilder code.
- I created a new distribution that included my changes to the gmbuilder code, compressed it, and uploaded it as a new version of the old distribution file: File:Dist cw20151201.zip. I then used the new version of GenMAPP Builder present in this file to run Trial II below.
- Trial 2- On the final step of the Export Wizard, the Bordetella pertussis species profile available was correctly listed as a custom profile. Therefore, I proceeded with the export. The testing report for this new gene database, labeled "cw20151201" can be found here: Gene_Database_Testing_Report-_cw20151201.
- Trial 1- On the final step of the Export Wizard, the Bordetella pertussis species profile available was listed as a generic profile. This demonstrated that the new version of GenMAPP Builder was not coded properly.
- Having verified that the GenMAPP Builder code worked as intended, I proceeded to commit my code to Github.
Committing the Changes to Github
- I right-clicked on the gmbuilder project folder and chose Team > Synchronize Workspace from the menu that appeared.
- Eclipse asked whether it is OK to enter the Team Synchronizing perspective, and I responded with Yes. I also checked the Remember my decision checkbox to prevent this from happening again.
- This switched me to the Team Synchronization perspective.
- I right-clicked on the gmbuilder project folder and chose Pull from the menu that appeared.
- I commited the updated files (marked with right-pointing gray-arrows) by right-clicking on them and choosing Commit....
- I checked to see if the files I wanted to commit were checked in the ensuing Commit Changes dialog. One of the two files was not, so I had to check it manually.
- I briefly described the nature of the changes that I was committing: "Created a new species profile within gmbuilder for b-pertussis".
- I chose Commit and Push.
Bordetella Pertussis Species Profile Customization
ID inconsistencies
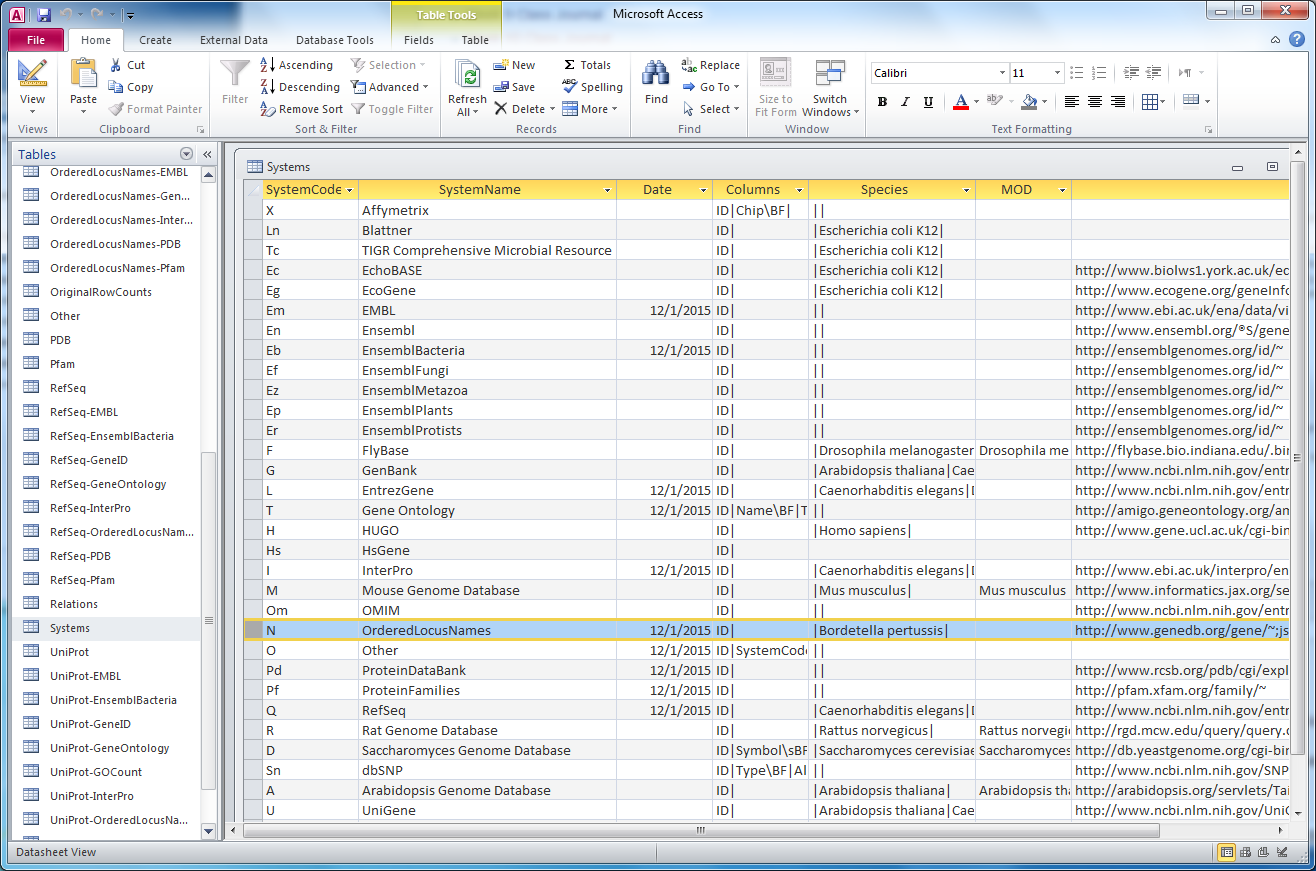
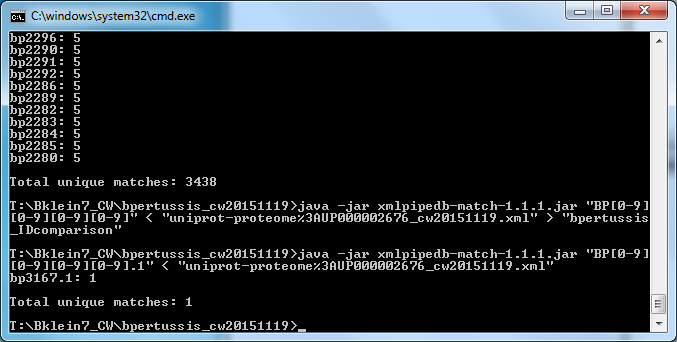
To assess the need for further customization of the Bordetella pertussis species profile, I worked with our QA specialist Mahrad to compare the gene IDs present in the cw20151201 .gdb file (File:Bpertussis-std cw20151201.zip) and the original .xml file (File:Uniprot-proteome-UP000002676 cw20151201.zip). This was done by copying the "OrderedLocusNames" table from the .gdb file into a new Excel document alongside the list of Bordetella pertussis gene IDs in the original .xml file that was generated using xmlpipedb match. Further details can be found in Mahrad's Week 14 Journal Entry. Overall, we found 4 discrepant IDs.
- BP3167.1- existed in the .gdb file but not in the the xmlpipedb match result
- Additionally, we found that the expected variant of this ID, BP3167, existed in the xmlpipedb match result but not in the .gdb file.
- We hypothesized that the ID existed as BP3167.1 in the original .xml file but was retrieved incompletely in the xmlpipedb match result due to the use of an imprecise regex (BP####).
- BP0101- did not get exported into the .gdb file
- To investigate why this gene was not exported, we looked up this ID in the original .xml file.
- We found that the gene ID was actually listed as both BP0101A and BP0101B, which was not retrieved in the xmlpipedb match result due to the use of an imprecise regex (BP####).
- The gene IDs BP0101A and BP0101B were both listed as open reading frame (ORF) genes in the .xml file as opposed to ordered locus names. This presented a new category of gene IDs that the custom Bordetella pertussis species profile was not designed to export.
- ORF investigation
- To see if there were other ORF genes with the pattern BP####A or BP####B, we ran a query to retrieve such IDs from the original PostgreSQL database: bpertussis_cw20151119_gmb3build5.
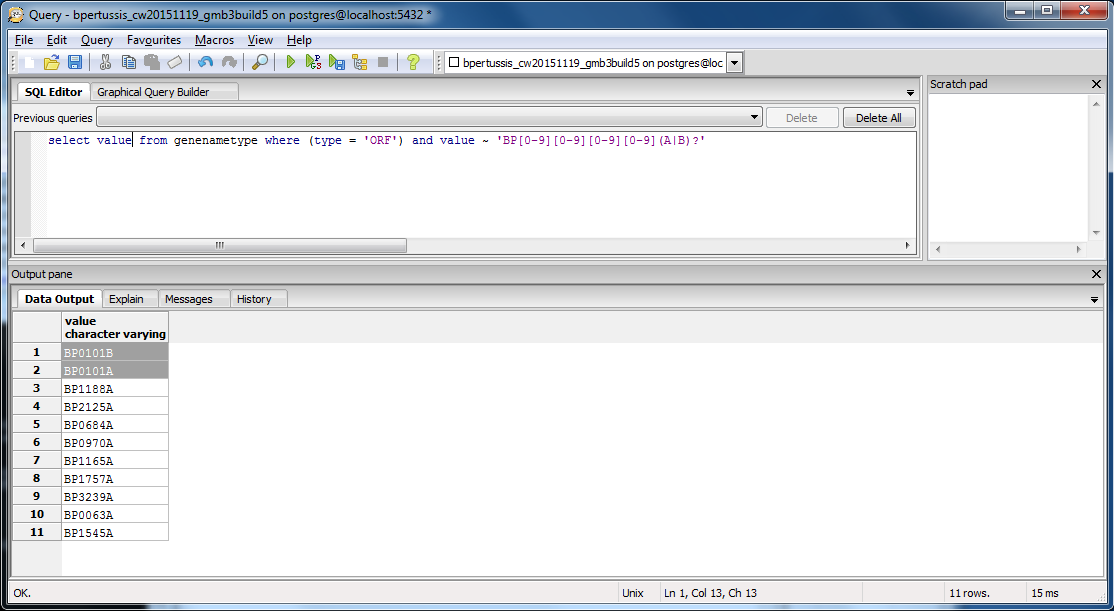
- This query retrieved a list of 11 ORF genes. Seven of these genes exhibited numerical sequences (####) that were cross-listed as ordered locus names, whereas four of the genes exhibited unique numerical sequences that were only listed as ORF genes. Therefore, only four of the ORF IDs should been discrepant during our comparative analysis between .gdb and .xml IDs. Of these four, two were BP0101A and BP0101B.
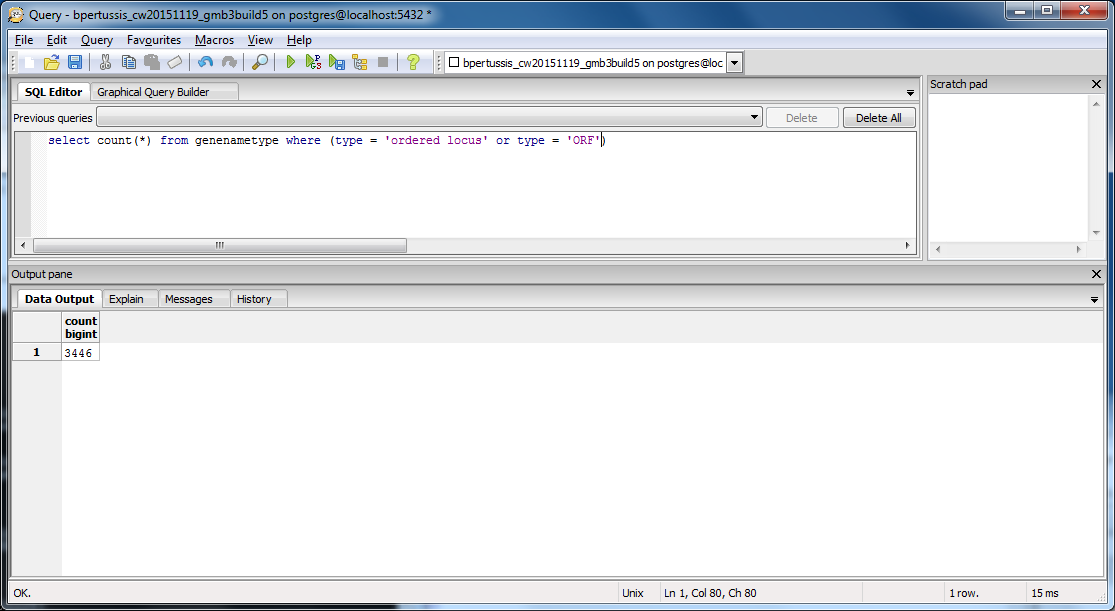
- To evaluate the total number of genes that should be exported into the .gdb file, including both genes listed under the ordered locus names and ORF tables, we queried the original PostgreSQL database once more:
- To see if there were other ORF genes with the pattern BP####A or BP####B, we ran a query to retrieve such IDs from the original PostgreSQL database: bpertussis_cw20151119_gmb3build5.
- To investigate why this gene was not exported, we looked up this ID in the original .xml file.
- BP0970- did not get exported into the .gdb file
- This was the third ORF gene with a unique numerical sequence. The gene was listed as BP0970A in the original .xml file.
- BP0684- did not get exported into the .gdb file
- This was the third ORF gene with a unique numerical sequence. The gene was listed as BP0684A in the original .xml file.
Conclusion: The custom Bordetella pertussis species profile in GenMAPP Builder needs to be customized to export the 11 ORF genes. The gene BP3167.1 was already being exported and, therefore, does not warrant coding changes. Instead, this unique pattern should be incorporated into the regex used to count Bordetella pertussis gene IDs when running future gene database testing reports.
Changes to the Code
To make changes to the GenMAPP Builder code, I opened the gmbuilder project within the Java perspective using the program Eclipse. The details regarding the developer rig I used can be found on my Week 12 Assignment Page.
- Adding a New Method Block to Import ORF Listings
- With the help of Dr. Dionisio, I added the following method block to the Bordetella pertussis custom species profile code:
@Override
public TableManager getSystemTableManagerCustomizations(TableManager tableManager,
TableManager primarySystemTableManager, Date version) throws SQLException, InvalidParameterException {
/*
* This method is only called (and therefore this bit 'o logic is only
* invoked) when the species specific class has not overridden this
* method.
*/
List<String> comparisonList = new ArrayList<String>(1);
comparisonList.add("ordered locus");
comparisonList.add("ORF");
return systemTableManagerCustomizationsHelper(tableManager, primarySystemTableManager, version,
"OrderedLocusNames", comparisonList);
}
- Committing and Pushing the New Code to Github
- I followed the steps outlined in the "Committing the Changes to Github" section of this journal entry to commit this updated code to Github.
- This version of GenMAPP Builder was labelled cw20151203.
- Compressing and Exporting the New Distribution Folder
- I followed the steps outlined in the "Creating a New Build of GenMAPP Builder" section of this journal entry to create a new GenMAPP Builder distribution folder and upload it to the wiki.
- cw20151203 distribution folder: File:Dist cw20151203.zip.
Running an Export with the New Build (cw20151203)
I downloaded GenMAPP Builder version cw20151203 onto the lab computer and ran a new export using the PostgreSQL database bpertussis_cw20151201_gmb3build5.
- Gene Database Testing Report: Gene Database Testing Report- cw20151203.
Links
- User Page: Brandon Klein
- Team Page: The Class Whoopers
Assignments Pages
- Week 1 Assignment
- Week 2 Assignment
- Week 3 Assignment
- Week 4 Assignment
- Week 5 Assignment
- Week 6 Assignment
- Week 7 Assignment
- Week 8 Assignment
- Week 9 Assignment
- Week 10 Assignment
- Week 11 Assignment
- Week 12 Assignment
- No Week 13 Assignment
- Week 14 Assignment
- Week 15 Assignment
Individual Journal Entries
- Week 1 Individual Journal
- Week 2 Individual Journal
- Week 3 Individual Journal
- Week 4 Individual Journal
- Week 5 Individual Journal
- Week 6 Individual Journal
- Week 7 Individual Journal
- Week 8 Individual Journal
- Week 9 Individual Journal
- Week 10 Individual Journal
- Week 11 Individual Journal
- Week 12 Individual Journal
- No Week 13 Journal
- Week 14 Individual Journal
- Week 15 Individual Journal
- Week 1 Class Journal
- Week 2 Class Journal
- Week 3 Class Journal
- Week 4 Class Journal
- Week 5 Class Journal
- Week 6 Class Journal
- Week 7 Class Journal
- Week 8 Class Journal
- Week 9 Class Journal
- Week 10 Team Journal
- Week 11 Team Journal
- Week 12 Team Journal
- No Week 13 Journal
- Week 14 Team Journal
- Week 15 Team Journal