Blitvak Week 6
Contents
Individual Journal Assignment Week 6
Downloading and Decompressing Data Files, Other Assignment Preparation
- Looking over the Week 6 Assignment Page, I found that
curl -O http://www.fda.gov/downloads/Drugs/InformationOnDrugs/UCM054599.zipcan be used, while I am in the /nfs/home/blitvak directory, to place a .zip containing the data files directly into my personal folder. I found thatunzip UCM054599.zipunzips the files into the personal folder. - I also downloaded and installed pgAdmin III from http://www.pgadmin.org/
- I spent some time reviewing the PostgreSQL Tutorial
- I booted up PuTTy and unzipped the files into my home folder
Working with application.txt
Opening and Reviewing the File
- I opened up and reviewed the application.txt and Product.txt files using
more <filename.txt>; I found that the data column labels for application.txt are:
ApplNo ApplType SponsorApplicant MostRecentLabelAvailableFlag CurrentPatentFlag ActionType Chemical_Type Ther_Potential Orphan_Code
- Reviewing the actual data, and with PostgreSQL in mind, I found that the variable type for each column should be:
ApplNo: int(primary key) ApplType: varchar SponsorApplicant: varchar MostRecentLabelAvailableFlag: boolean CurrentPatentFlag: boolean ActionType: varchar Chemical_Type: int Ther_Potential: varchar Orphan_Code: varchar
- I realized that any empty data spaces in application.txt will have to be turned into
null - I realized that
sed "1D"will have to be executed in order to remove the first row (which is column labeling) - Referencing the Week 6 Assignment Page, I learned that the data within these text files is separated by tabs (
\t) instead of commas
Modifying application.txt
- I opened the file and decided to try to turn the tabs into commas using
cat application.txt | sed "s/\t/,/g"- Adding onto that, I decided to get rid of the other spaces between the data by using
sed -e "s/\s\{4,\}//g", which matches 4 whitespaces and removes them. This command was found in a StackExchange post.
- Adding onto that, I decided to get rid of the other spaces between the data by using
- At this point, I noticed that many lines had extra commas either at the ends of the lines or in the middle, indicating missing or nonexistent values
- I used
sed "s/,,,\r$/,null,null,null/;s/,,\r$/,null,null/;s/,\r$/,null/1" | sed "s/,,/,null/;s/,,,/,null,null/g", along with what I already have, in order to turn any extra commas (missing values) into null
- I used
- My focus now turned to making sure that all data that was identified as being varchar has apostrophes wrapping around it.
- Working with the ApplType data and my previous work, I used
sed "s/......,/&'/1" | sed "s/'./&'/1"to surround it with apostrophes. - Working with the SponsorApplicant data, I added
sed "s/',/&'/g" | sed "s/,./'&/3"to wrap apostrophes around the third data type in each line - I used
sed "s/,/&'/5" | sed "s/'../&'/4", to place apostrophes around the two character data under ActionType
- Working with the ApplType data and my previous work, I used
- Using
grep "nullP",grep "nullS", andgrep "nullV"with the current pipeline of commands:cat application.txt | sed "1D" | sed "s/\t/,/g" | sed -e "s/\s\{4,\}//g" | sed "s/,,,\r$/,null,null,null/;s/,,\r$/,null,null/;s/,\r$/,null/1" | sed "s/,,/,null/;s/,,,/,null,null/g" | sed "s/......,/&'/1" | sed "s/'./&'/1" | sed "s/',/&'/g" | sed "s/,./'&/3" | sed "s/,/&'/5" | sed "s/'../&'/4"- I noticed that some of the null values are not separated from adjacent data with commas; I added
sed "s/nullS/null,S/g" | sed "s/nullV/null,V/g" | sed "s/nullP/null,P/g"to fix this issue and I checked the result usinggrep - With the Ther_Potential data being now completely separated by commas from the other data, I then proceeded to try to surround it with apostrophes. I first added
sed "s/,'..',.,/&'/g"to add the first apostrophe, and I noticed that this command led some null values to gain an apostrophe. I addedsed "s/'null/null/g"/<code> to clean them up. I later noticed that some Ther_potential values have asterisks tied to them, I used <code>grep "P\*"andgrep "S\*"to confirm the presence of asterisks. Finally, I addedsed "s/'S,null/'S',null/g" | sed "s/'P,null/'P',null/g" | sed "s/'S,V/'S',V/g" | sed "s/'P,V/'P',V/g" | sed "s/'P\*/'P\*'/g" | sed "s/'S\*/'S\*'/g"to the pipeline to fully surround the Ther_Potential values. - I surrounded the Orphan_Code variable with apostrophes by adding
sed "s/,V/,'V'/g"to the pipeline (Orphan_Code is often null but when it is present, it is always a V)
- I finished formatting the file by adding
sed "s/^/insert into applications(ApplNo,ApplType,SponsorApplicant,MostRecentLabelAvailableFlag,CurrentPatentFlag,ActionType,Chemical_Type,Ther_Potential,Orphan_Code) values(/g" | rev | sed -r "s/llun|'V'/;)&/1" | revto the pipeline. I decided to reverse the file in this set of commands because I could not getsedto add ); to the very end of each line; I decided upon creating a multiple choicesedcommand that worked on the very first match of a line.
Generating the application.sql.txt file
- I generated the application.sql.txt file using
cat application.txt | sed "1D" | sed "s/\t/,/g" | sed -e "s/\s\{4,\}//g" | sed "s/,,,\r$/,null,null,null/;s/,,\r$/,null,null/;s/,\r$/,null/1" | sed "s/,,/,null/;s/,,,/,null,null/g" | sed "s/......,/&'/1" | sed "s/'./&'/1" | sed "s/',/&'/g" | sed "s/,./'&/3" | sed "s/,/&'/5" | sed "s/'../&'/4" | sed "s/nullS/null,S/g" | sed "s/nullV/null,V/g" | sed "s/nullP/null,P/g" | sed "s/,'..',.,/&'/g" | sed "s/'null/null/g" | sed "s/'S,null/'S',null/g" | sed "s/'P,null/'P',null/g" | sed "s/'S,V/'S',V/g" | sed "s/'P,V/'P',V/g" | sed "s/'P\*/'P\*'/g" | sed "s/'S\*/'S\*'/g" | sed "s/,V/,'V'/g" | sed "s/^/insert into applications(ApplNo,ApplType,SponsorApplicant,MostRecentLabelAvailableFlag,CurrentPatentFlag,ActionType,Chemical_Type,Ther_Potential,Orphan_Code) values(/g" | rev | sed -r "s/llun|'V'/;)&/1" | rev > ~/public_html/application.sql.txt - I proceeded to input
http://my.cs.lmu.edu/~blitvak/application.sql.txtinto my browser and I copied all of the data
Testing application.sql.txt in pgAdmin III
- I created a table in postgreSQL using
create table applications (ApplNo int primary key, ApplType varchar, SponsorApplicant varchar, MostRecentLabelAvailableFlag boolean, CurrentPatentFlag boolean, ActionType varchar, Chemical_Type int, Ther_Potential varchar, Orphan_Code varchar) - I pasted the data into the program and noticed these types of errors:
1. Type 1
ERROR: syntax error at or near "lse" LINE 17882: ... values(125422,'B','THROMBOGENICS', INC,False,'Fa'lse,AP,nul...
2. Type 2
ERROR: column "s" does not exist LINE 161: ...e) values(008319,'N','NOVARTIS',False,False,'AP',14,S,null); ERROR: column "s" does not exist LINE 360: ...10721,'N','CITRON PHARMA LLC',False,False,'AP',null,S,null); ERROR: column "p" does not exist LINE 617: ...de) values(013025,'N','HOSPIRA',False,False,'AP',14,P,null);
- The Type 1 Error, I noticed was linked to the presence of commas in the original raw data with respect to the many company names, should be fixed by initially replacing the commas with a character that does not exist in the data, such as ~;
sed "s/,/~/g"was added near the beginning of the pipe, andsed "s/~/,/g"was added at the end of the pipe. - The Type 2 Error seems to be tied to the presence of a double digit Chemical_Type; it might be remedied by adding
sed "s/,S,/,'S',/;s/,P,/,'P',/;s/,S\*,/,'S\*',/;s/,P\*,/,'P\*',/g"near the end of the pipe.- The modified pipe to be tested again is:
cat application.txt | sed "s/,/~/g" | sed "1D" | sed "s/\t/,/g" | sed -e "s/\s\{4,\}//g" | sed "s/,,,\r$/,null,null,null/;s/,,\r$/,null,null/;s/,\r$/,null/1" | sed "s/,,/,null/;s/,,,/,null,null/g" | sed "s/......,/&'/1" | sed "s/'./&'/1" | sed "s/',/&'/g" | sed "s/,./'&/3" | sed "s/,/&'/5" | sed "s/'../&'/4" | sed "s/nullS/null,S/g" | sed "s/nullV/null,V/g" | sed "s/nullP/null,P/g" | sed "s/,'..',.,/&'/g" | sed "s/'null/null/g" | sed "s/'S,null/'S',null/g" | sed "s/'P,null/'P',null/g" | sed "s/'S,V/'S',V/g" | sed "s/'P,V/'P',V/g" | sed "s/'P\*/'P\*'/g" | sed "s/'S\*/'S\*'/g" | sed "s/,V/,'V'/g" | sed "s/^/insert into applications(ApplNo,ApplType,SponsorApplicant,MostRecentLabelAvailableFlag,CurrentPatentFlag,ActionType,Chemical_Type,Ther_Potential,Orphan_Code) values(/g" | rev | sed -r "s/llun|'V'/;)&/1" | rev | sed "s/,S,/,'S',/;s/,P,/,'P',/;s/,S\*,/,'S\*',/;s/,P\*,/,'P\*',/g" | sed "s/~/,/g"
- This new pipeline revealed another error that I did not notice earlier (but resulted in the fixing of the previous errors):
ERROR: INSERT has more target columns than expressions LINE 4864: ...atentFlag,ActionType,Chemical_Type,Ther_Potential,Orphan_Cod... insert into applications(ApplNo,ApplType,SponsorApplicant,MostRecentLabelAvailableFlag,CurrentPatentFlag,ActionType,Chemical_Type,Ther_Potential,Orphan_Code) values(125057,'B','ABBVIE INC',False,False,'AP',null,'V');
- It seems that every line with a null instead of Chemical_Type value, prior to an Orphan_Code, is missing a null value (common pattern is
'AP',null,'V')- Inserting
sed "s/'AP',null,'V'/'AP',null,null,'V'/g"near the end of the pipe might correct this issue.
- Inserting
- The fix worked and the final pipe resulted in a successful table creation. The final pipeline of commands is:
cat application.txt | sed "s/,/~/g" | sed "1D" | sed "s/\t/,/g" | sed -e "s/\s\{4,\}//g" | sed "s/,,,\r$/,null,null,null/;s/,,\r$/,null,null/;s/,\r$/,null/1" | sed "s/,,/,null/;s/,,,/,null,null/g" | sed "s/......,/&'/1" | sed "s/'./&'/1" | sed "s/',/&'/g" | sed "s/,./'&/3" | sed "s/,/&'/5" | sed "s/'../&'/4" | sed "s/nullS/null,S/g" | sed "s/nullV/null,V/g" | sed "s/nullP/null,P/g" | sed "s/,'..',.,/&'/g" | sed "s/'null/null/g" | sed "s/'S,null/'S',null/g" | sed "s/'P,null/'P',null/g" | sed "s/'S,V/'S',V/g" | sed "s/'P,V/'P',V/g" | sed "s/'P\*/'P\*'/g" | sed "s/'S\*/'S\*'/g" | sed "s/,V/,'V'/g" | sed "s/^/insert into applications(ApplNo,ApplType,SponsorApplicant,MostRecentLabelAvailableFlag,CurrentPatentFlag,ActionType,Chemical_Type,Ther_Potential,Orphan_Code) values(/g" | rev | sed -r "s/llun|'V'/;)&/1" | rev | sed "s/,S,/,'S',/;s/,P,/,'P',/;s/,S\*,/,'S\*',/;s/,P\*,/,'P\*',/g" | sed "s/~/,/g" | sed "s/'AP',null,'V'/'AP',null,null,'V'/g" > ~/public_html/application.sql.txt
- I replaced the previously made application.sql.txt with the output of the final pipeline: Output
Working with Product.txt
Opening and Reviewing Product.txt
- Product.txt was examined using
more Product.txtand the column labels and variable type were found to be:
ApplNo(int) ProductNo(int) Form(varchar) Dosage(varchar) ProductMktStatus(int) TECode(varchar) ReferenceDrug(int) drugname(varchar) activeingred(varchar)
Modifying Product.txt
- I removed the first row (column labels) and replaced all of the tabs with commas; I also turned the extra commas into null values:
cat Product.txt | sed "1D" | sed "s/\t/,/g" | sed "s/,,/,null,/g" - I began placing the apostrophes around the varchar data with the Form, Dosage, and TECode data. I placed apostrophes around this data by adding several
sedreplacements to the pipeline made earlier:sed "s/,/&'/2" | sed "s/,/'&/3" | sed "s/,/&'/3" | sed "s/,/'&/4" | sed "s/,/&'/5" | sed "s/,/'&/6" - I noticed that there are a few commas and apostrophes in the raw data, I decided to initially replace the commas with characters that are not present elsewhere in the raw data (such as~);
sed "s/,/~/g"was added to the beginning of the command chain to initially replace them, andsed "s/~/,/g"was added to the end of chain in order to restore them. The presence of apostrophes in the data was fixed by through addingsed "s/'/''/g"to the beginning of the pipe (this addition allows pgSQL to understand that these apostrophes are a part of the actual data. - I placed apostrophes around all remaining varchar data by adding
sed "s/,/&'/7" | sed "s/,/'&/8" | sed "s/,/&'/8" | sed "s/\r$/'/g"to the pipeline - To the end of the current pipeline, I added
sed "s/^/insert into products(ApplNo,ProductNo,Form,Dosage,ProductMktStatus,TECode,ReferenceDrug,drugname,activeingred) values(/g" | sed "s/$/);/g"to wrap up the formatting of the raw data (should now be usable as sequences of SQLinsertstatements) - I used the final pipeline of commands:
cat Product.txt | sed "1D" | sed "s/,/~/g" | sed "s/'/''/g" | sed "s/\t/,/g" | sed "s/,,/,null,/g" | sed "s/,/&'/2" | sed "s/,/'&/3" | sed "s/,/&'/3" | sed "s/,/'&/4" | sed "s/,/&'/5" | sed "s/,/'&/6" | sed "s/,/&'/7" | sed "s/,/'&/8" | sed "s/,/&'/8" | sed "s/\r$/'/g" | sed "s/^/insert into products(ApplNo,ProductNo,Form,Dosage,ProductMktStatus,TECode,ReferenceDrug,drugname,activeingred) values(/g" | sed "s/~/,/g" | sed "s/$/);/g" > ~/public_html/Product.sql.txtto generate Product.sql.txt
Testing Products.sql.txt in pgAdmin III
- The table for the data enclosed in Products.sql.txt was produced using
create table products (ApplNo int, ProductNo int, Form varchar, Dosage varchar, ProductMktStatus int, TECode varchar, ReferenceDrug int, drugname varchar, activeingred varchar)). The contents of this file was checked in pgAdmin III/pgSQL with no issues.
Review/Questions to Answer
- Provide the DDL (
create table) statements that you used for your application and product tables.- For the applications table:
create table applications (ApplNo int primary key, ApplType varchar, SponsorApplicant varchar, MostRecentLabelAvailableFlag boolean, CurrentPatentFlag boolean, ActionType varchar, Chemical_Type int, Ther_Potential varchar, Orphan_Code varchar) - For the products table:
create table products (ApplNo int, ProductNo int, Form varchar, Dosage varchar, ProductMktStatus int, TECode varchar, ReferenceDrug int, drugname varchar, activeingred varchar)
- For the applications table:
- Provide the
sedcommand sequences that you used to convert the raw text files into sequences of SQLinsertstatements.- For the application.txt data:
cat application.txt | sed "s/,/~/g" | sed "1D" | sed "s/\t/,/g" | sed -e "s/\s\{4,\}//g" | sed "s/,,,\r$/,null,null,null/;s/,,\r$/,null,null/;s/,\r$/,null/1" | sed "s/,,/,null/;s/,,,/,null,null/g" | sed "s/......,/&'/1" | sed "s/'./&'/1" | sed "s/',/&'/g" | sed "s/,./'&/3" | sed "s/,/&'/5" | sed "s/'../&'/4" | sed "s/nullS/null,S/g" | sed "s/nullV/null,V/g" | sed "s/nullP/null,P/g" | sed "s/,'..',.,/&'/g" | sed "s/'null/null/g" | sed "s/'S,null/'S',null/g" | sed "s/'P,null/'P',null/g" | sed "s/'S,V/'S',V/g" | sed "s/'P,V/'P',V/g" | sed "s/'P\*/'P\*'/g" | sed "s/'S\*/'S\*'/g" | sed "s/,V/,'V'/g" | sed "s/^/insert into applications(ApplNo,ApplType,SponsorApplicant,MostRecentLabelAvailableFlag,CurrentPatentFlag,ActionType,Chemical_Type,Ther_Potential,Orphan_Code) values(/g" | rev | sed -r "s/llun|'V'/;)&/1" | rev | sed "s/,S,/,'S',/;s/,P,/,'P',/;s/,S\*,/,'S\*',/;s/,P\*,/,'P\*',/g" | sed "s/~/,/g" | sed "s/'AP',null,'V'/'AP',null,null,'V'/g" > ~/public_html/application.sql.txt - For the Product.txt data:
cat Product.txt | sed "1D" | sed "s/,/~/g" | sed "s/'/''/g" | sed "s/\t/,/g" | sed "s/,,/,null,/g" | sed "s/,/&'/2" | sed "s/,/'&/3" | sed "s/,/&'/3" | sed "s/,/'&/4" | sed "s/,/&'/5" | sed "s/,/'&/6" | sed "s/,/&'/7" | sed "s/,/'&/8" | sed "s/,/&'/8" | sed "s/\r$/'/g" | sed "s/^/insert into products(ApplNo,ProductNo,Form,Dosage,ProductMktStatus,TECode,ReferenceDrug,drugname,activeingred) values(/g" | sed "s/~/,/g" | sed "s/$/);/g" > ~/public_html/Product.sql.txt
- For the application.txt data:
- Using the command line, how can you determine the number of records in each file? Provide the command.
- One can use
wc <file_name>(where<file_name>could be application.txt or Product.txt) to determine the number of records in each file; the output ofwcis in the format (from left to right) of lines, words, characters; for these two files, since each entry occupies its own line and since the first line is just column labeling, the number of records would be the lines output ofwc <file_name>minus one. - For application.txt, the command is
wc application.txtwith an output of: 19747 147336 1615694 application.txt (this corresponds to 19746 records) - For Product.txt, the command is
wc Product.txtwith an output of: 32771 369379 2856017 Product.txt (this corresponds to 32770 records)
- One can use
- Using SQL, how can you determine the number of records in the table corresponding to the file? Provide the SQL select statement.
- For applications table:
select count(*) from applications, output of 19746 entries - For products table:
select count(*) from products, output of 32770 entries
- For applications table:
- In your database, are these numbers the same or different? Explain why you think so.
- The numbers as shown in the database represent the same number of records as those given by the command line command (a user will just need to take into account the fact that there exists a line for column labels in the original text files; this line should not be counted as a record, and therefore, the number of records needs to be subtracted by one with respect to the command line output for lines. The column label line was deleted when the data was formatted for use by pgSQL, and thus, the output number of records in the database (for each data set) is one less from that of the command line output (which is the lines section of the
wccommand).
- The numbers as shown in the database represent the same number of records as those given by the command line command (a user will just need to take into account the fact that there exists a line for column labels in the original text files; this line should not be counted as a record, and therefore, the number of records needs to be subtracted by one with respect to the command line output for lines. The column label line was deleted when the data was formatted for use by pgSQL, and thus, the output number of records in the database (for each data set) is one less from that of the command line output (which is the lines section of the
- What are the names of the drug products that are administered in the form
INJECTABLE;INTRAVENOUS, SUBCUTANEOUS?- The
drugnamevariable represents the names of the drug products; theFormvariable represents the form that the drug is administered in. - The command to find out this information is:
select drugname from products where form = 'INJECTABLE;INTRAVENOUS, SUBCUTANEOUS' - The names of the drug products are:LOVENOX, VELCADE, VIDAZA, ENOXAPARIN SODIUM, ACTEMRA, and AZACITIDINE. (one entry for each drug product)
- The
- What are the names of the drug products whose active ingredient (activeingred) is
ATROPINE?- The command to find out this information is:
select drugname from products where activeingred = 'ATROPINE' - The names of the drug products are: ATROPEN (four entries) and ATROPINE (one entry)
- The command to find out this information is:
- In what forms and dosages can the drug product named
BENADRYLbe administered?- The command to find out this information is:
select Form, Dosage from products where drugname = 'BENADRYL' - The forms that
BENADRYLcan be administered in, include: CAPSULE;ORAL, ELIXIR;ORAL, and INJECTABLE;INJECTION - The dosages that
BENADRYLcan be administered in, include: 50MG, 12.5MG/5ML, 25MG, 10MG/ML, and 50MG/ML.
- The command to find out this information is:
- Which drug products have a name ending in
ESTROL?- Reading over the PostgreSQL Tutorial, I learned that % is interpreted as a wildcard by pgSQL
- The command to find out this information is:
select drugname from products where drugname like '%ESTROL' - Drug products that have a name ending in
ESTROL: DIETHYLSTILBESTROL, STILBESTROL, DIENESTROL
- Produce a table listing all of the known values for the therapeutic_potential column in the application table and how many application records there are of each
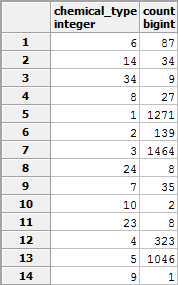
- Produce a table listing all of the known values for the chemical_type column in the application table and how many application records there are of each.
- What are the names of the drug products that are sponsored (sponsor applicant column) by MERCK?
- Outline:
select columns from table1 inner join table2 on (join condition) where conditions, taken from the PostgreSQL Tutorial - Command:
select drugname from products inner join applications on (products.ApplNo = applications.ApplNo) where SponsorApplicant = 'MERCK' group by drugname - Names: DECADRON, HUMORSOL, NEO-HYDELTRASOL, PRINIVIL, MAXALT-MLT, HYDROCORTONE, PERIACTIN, PROPECIA, PROSCAR, CLINORIL, PRINZIDE, ELSPAR, ALDOMET, ALDORIL D30, EMEND, DIUPRES-500, NEODECADRON, ALDOCLOR-250, MEFOXIN IN DEXTROSE 5% IN PLASTIC CONTAINER, ARAMINE, TIAMATE, BLOCADREN, CANCIDAS, CHIBROXIN, CORTONE, PEPCID, TRUSOPT, REDISOL, VIOXX, FLOROPRYL, ALPHAREDISOL, DECADRON-LA, MEFOXIN IN SODIUM CHLORIDE 0.9% IN PLASTIC CONTAINER, PEPCID RPD, NOROXIN, ZOCOR, LERITINE, DECADERM, DOLOBID, MANNITOL 25%, DECADRON W/ XYLOCAINE, ALDORIL D50, TIMOLIDE 10-25, CYCLAINE, HYDROPRES 25, AMINOHIPPURATE SODIUM, MEVACOR, MODURETIC 5-50, ALDORIL 25, SINGULAIR, COLBENEMID, DIUPRES-250, HYDELTRA-TBA, PRIMAXIN, BENEMID, MAXALT, FOSAMAX PLUS D, HYDRODIURIL, HYDELTRASOL, HYDROPRES 50, ZOLINZA, ALDORIL 15, FOSAMAX, DECASPRAY, COGENTIN, ALDOCLOR-150, PEPCID PRESERVATIVE FREE
- Outline:
- Which sponsor applicant companies have the text LABS in their names and have products whose active ingredients (activeingred) include both
ASPIRINandCAFFEINE?- Command:
select SponsorApplicant from applications inner join products on (applications.ApplNo = products.ApplNo) where SponsorApplicant like '%LABS%' and activeingred like '%ASPIRIN%' and activeingred like '%CAFFEINE%' group by SponsorApplicant - ACTAVIS LABS UT INC and WATSON LABS
- Command:
Brandon Litvak
BIOL 367, Fall 2015
| Weekly Assignments | Individual Journal Pages | Shared Journal Pages |
|---|---|---|
|
|
|
|