Jkuroda Week 11

Contents
Preparation for Journal Club
Definitions
- Bioremediation: use of biological organisms to solve an environmental problem such as contaminated soil or groundwater. Source
- Orthologous: two genes are to be orthologous if they diverged after a speciation event. Source
- Lysogeny: the fusion of the nucleic acid of a bacteriophage with that of a host bacterium so that the potential exists for the newly integrated genetic material to be transmitted to daughter cells at each subsequent cell division. Source
- Reductase: an enzyme that catalyzes a chemical reduction. Source
- Translocation: a chromosomal segment is moved from one position to another, either within the same chromosome or to another chromosome. Source
- Deamination: removal of the amino group, -NH2, from a compound. Source
- Fumarate: a dicarboxylic acid intermediate in the tricarboxylic acid cycle. Source
- Biofilm: an aggregate of microbes with a distinct architecture. A biofilm is like a tiny city in which microbial cells, each only a micrometer or two long, form towers that can be hundreds of micrometers high. Source
- Plasmid: a linear or circular double-stranded DNA that is capable of replicating independently of the chromosomal DNA. Source
- Intergenic Regions: DNA sequences located between genes that comprise a large percentage of the human genome with no known function. Source
Outline
Significance of this work
- Shewanella oneidensis possesses diverse respiratory capabilities, which is partially enabled by multi-component/branched electron transport systems.
- This makes shewanella oneidensis a significant model organism for bioremediation studies, which deal with solutions to environmental issues like contaminated soil or groundwater.
Methods used in the study
- The study sequenced the shewanella oneidensis genome using the whole-genome sequencing method.
Results
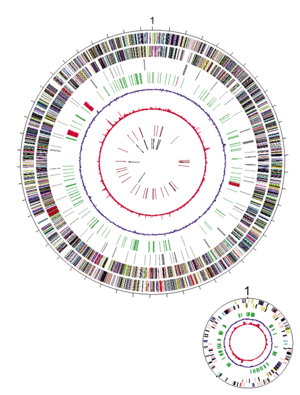
The shewanella oneidensis genome is a circular chromosome of exactly 4,969,803 base pairs. A total of 4,758 predicted protein-encoding open reading frames are present and of those, 23.4% are unique to this bacterium.
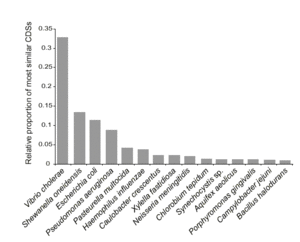
Most of the shewanella oneidensis MR-1 predicted protein-encoding open reading frames are similar to vibrio cholerae genes. Shwanella oneidensis MR-1 matched with 1,265 predicted protein-encoding open reading frames in vibrio cholerae, 32.33% of the vibrio cholerae genome. Furthermore, 683 of the shewanella oneidensis predicted protein-encoding open reading frames are similar to other shewanella oneidensis genes, which suggests lineage-specific duplications.
The study revealed that vibrio cholerae was the only organism with extensive regions of similar gene order. These syntenic regions are only on the vibrio cholerae chromosome I. This suggests that the second chromosome of vibrio cholerae was captured after the divergence of vibrio cholerae and shewanella oneidensis, that the second chromosome was lost in the shewanella oneidensis lineage, or that the second chromosome was rearranged in the shewanella lineage.
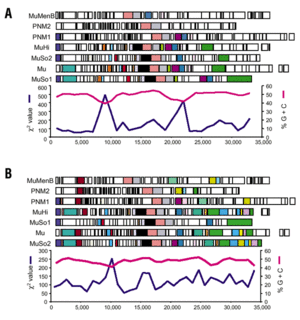
Genome analysis revealed a 51,857 base-pair lambda-like phage genome, both integrated in the shewanella oneidensis genome and present in non-integrated form, suggesting that it is a functional phage. The discovery of the shewanella lambda-like phage may provide an avenue for genetic manipulation of this group of microbes and allow the design of strains for specific bioremediation purposes.
Comparison to previous studies
- Through genome analysis, 39 c-type cytochromes were revealed, including 32 previously unidentified in shewanella oneidensis, and a novel periplasmic hydrogenase, integral members of the electron transport system.
- Shewanella oneidensis was found to possess a classic hydrogenase operon and a heterodimeric hydrogenase, which is an important element for its anaerobic respiratory capability as well as its metal-reducing capability. There have been no previous reports of the presence of this hydrogenase enzyme family in a facultative aerobe.
Model Organism Database
- What types of data can be found in the database (sequence, structures, annotations, etc.); is it a primary or “meta” database; is it curated electronically, manually [in-house], or manually [community])?
- According to the database, there are protein-coding and non-coding genes, splice variants, cDNA and protein sequences, non-coding RNAs, gene families based on HAMAP and PANTHER classification, homologues and gene trees including species across the pan-taxonomic range. Because it cites a provider (European Nucleotide Archive), this is a "meta" database. The database from the ENA was last updated on November 16, 2012. It seems that data is curated both electronically and manually in-house. "Data submitted to the ENA are validated by automated quality checking and, where possible, manual inspection and curation."[1]
- What individual or organization maintains the database?
- "The ENA is developed and maintained at the EMBL-EBI under the guidance of the INSDC International Advisory Committee and a Scientific Advisory Board."[2]
- What is their funding source(s)?
- "ENA is developed and operated under the support of the European Molecular Biology Laboratory (EMBL) [33] and through grants from external bodies that include the Seventh Framework Programme of the European Commission [34] (EC-FP7), the British Biotechnology and Biological Sciences Research Council (BBSRC) [35] and the Wellcome Trust (WT)"[3]
- Is there a license agreement or any restrictions on access to the database?
- No, the database is open to the public.
- How often is the database updated?
- Although the website from which the database is accessed is updated every two months, the last update to the actual database was November 16, 2012.
- Are there links to other databases?
- Yes, because this website simply uses data from the ENA.
- Can the information be downloaded?
- Yes, in FASTA and GFF3 formats.
- Evaluate the “user-friendliness” of the database.
- Is the Web site well-organized?
- Yes, although it does seem to be one component of a multi-faceted system for accessing the genome database of many types of bacteria.
- Does it have a help section or tutorial?
- There is a help section, but it is for general website use, and it is not specifically meant for the shewanella oneidensis database.
- Run a sample query. Do the results make sense?
- I ran a search for
SO_0001. The results came back with a name, description, gene ID, species, location and synonyms. The results make sense to me.
- I ran a search for
- Is the Web site well-organized?
- What is the format (regular expression) of the main type of gene ID for this species (the "ordered locus name" ID)? (for example, for Vibrio cholerae it was VC#### or VC_####).
-
SO_####
-
Individual Journal Entries
- Week 2
- Week 3
- Week 4
- Week 5
- Week 6
- Week 7
- Week 8
- Week 9
- Week 10
- Week 11
- Week 12
- Week 13
- Week 14
- Week 15