Jkuroda Week 9
Contents
Export Information
Version of GenMAPP Builder: 3 build 5
Computer on which export was run: HP Compaq 8300 Elite SFF FC
Postgres Database name: Vibrio Cholerae_10-27-15_GMB3-build-5
UniProt XML filename: uniprot-organism%3A243277
- UniProt XML version: UniProt release 2015_10 - October 14, 2015
- UniProt XML download link
- Time taken to import: 3.14 minutes
- Note: n/a
GO OBO-XML filename: go daily-termdb.obo-xml
- GO OBO-XML version (The version information can be found in the file properties after the file downloaded from the GO Download page has been unzipped):
- GO OBO-XML download link
- Time taken to import: 7.46 minutes
- Time taken to process: 4.34 minutes
- Note: n/a
GOA filename: 46.V_cholerae_ATCC_39315.goa
- GOA version (News on this page records past releases; current information can be found in the Last modified field on the FTP site): October 14, 2015 - GOA Proteome Sets 124
- GOA download link
- Time taken to import: 0.07 minutes
- Note: n/a
Name of .gdb file: Vc-Std_20151027_JK.gdb
- Time taken to export: 1 hour and 15 minutes
- Start time: 3:55pm
- End time: 5:10pm
- Note: n/a
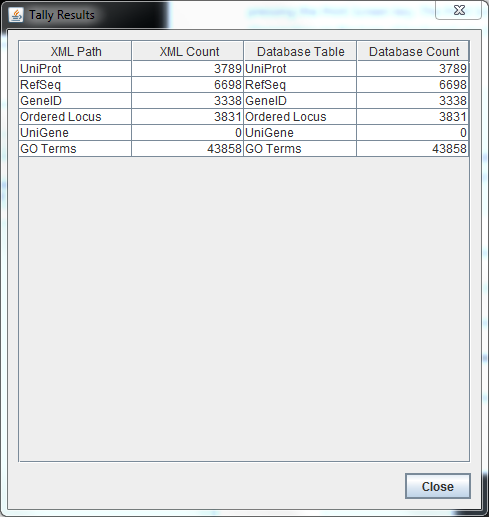
TallyEngine
- Run the TallyEngine in GenMAPP Builder and record the number of records for UniProt and GO in the XML data and in the Postgres databases.
- Choose the menu item Tallies > Run XML and Database Tallies for UniProt and GO...
- Take a screenshot of the results. Upload the image to the wiki and display it on this page.
- For more information, see this page.
Using XMLPipeDB match to Validate the XML Results from the TallyEngine
Follow the instructions found on this page to run XMLPipeDB match.
Are your results the same as you got for the TallyEngine? Why or why not?
- Initially, we got 3831 IDs for both XML and Postgres DB from TallyEngine but got 2738 IDs by using XMLPipeDB match. However, we realized that we were not accounting for the IDs that had the prefix of "VC_A" so we changed the command to reflect that:
java -jar xmlpipedb-match-1.1.1.jar "VC_A?[0-9][0-9][0-9][0-9]" < uniprot-organism%3A243277.xml
Once I had used the correct command, XMLPipeDB gave me a count of 3831 IDs.
Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine
For more information, see this page.
You can also look for counts at the SQL level, using some variation of a select count(*) query. This requires some knowledge of which table received what data. Here’s an initial tip: the gene/name tags in the XML file land in the genenametype table. A query on this table counting values from this table that were marked as ordered locus in the XML file matching the pattern VC_A?[0-9][0-9][0-9][0-9] would look like this:
select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_A?[0-9][0-9][0-9][0-9]';
In pgAdmin III, you can issue these queries by clicking on the pencil/SQL icon in the toolbar, typing the query into the SQL Editor tab, then clicking on the green triangular Play button to run.
Are your results the same as reported by the TallyEngine? Why or why not?
- Once again, we first got 2737 IDs by using SQL, which differed from the 3831 IDs from TallyEngine. But this was a result of the same mistake we made with XMLPipeDB Match. Once we accounted for the "VC_A" prefix, we got 3831 IDs.
OriginalRowCounts Comparison
Within the .gdb file, look at the OriginalRowCounts table to see if the database has the expected tables with the expected number of records. Compare the tables and records with a benchmark .gdb file.
Benchmark .gdb file: 2010 benchmark file
Copy the OriginalRowCounts table from the benchmark and new gdb and paste them here:
2010 Benchmark Original Row Counts Table
See Analysis section for more on the comparison and the discrepancy found because of this comparison.
Note: Using Microsoft Access, we found 7664 IDs, which was actually double the number of IDs present because of duplicated IDs that did not have an underscore.
Visual Inspection
Perform visual inspection of individual tables to see if there are any problems.
- Look at the Systems table. Is there a date in the Date field for all gene ID systems present in the database?
- No. For the current version, a good number of gene ID systems in the database do not have a value for the date field. Some systems that lack a date include: GenBank, UniGene, WormBase, and EcoGene.
- In the 2010 version, there are also missing dates. The systems listed above do not have a date in this version either.
- Open the UniProt, RefSeq, and OrderedLocusNames tables. Scroll down through the table. Do all of the IDs look like they take the correct form for that type of ID?
- For the UniProt tables, both versions seem have ID forms where most start with Q9. For the RefSeq tables, the 2010 version has ID forms that start with NP but my version has forms that start with either NP or WP. For the OrderedLocusNames tables, both versions seem to have the same ID form that either starts with
VCorVC_A. Notably, both versions have duplicates because of the removal of the underscore from the gene ID.
- For the UniProt tables, both versions seem have ID forms where most start with Q9. For the RefSeq tables, the 2010 version has ID forms that start with NP but my version has forms that start with either NP or WP. For the OrderedLocusNames tables, both versions seem to have the same ID form that either starts with
Note: n/a
Analysis
Consolidating the counts of gene IDs from the various methods, I got:
- 3831 IDs from Tally Engine
- 3831 IDs from xmlpipedb-match
java -jar xmlpipedb-match-1.1.1.jar "VC_A?[0-9][0-9][0-9][0-9]" < uniprot-organism%3A243277.xml
- 3831 IDs from PostgreSQL
select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_A?[0-9][0-9][0-9][0-9]';
- 3832 IDs from Microsoft Access (counting the IDs with an underscore)
Notice that there is a small but significant discrepancy in that there seems to be one more ID when we used Microsoft Access. This is troubling because of the fact that the other three methods seemed to confirm a total count of 3831. So, I used Microsoft Excel to compare the list of gene IDs from the actual .gdb file and the list I got back from PostgreSQL. As you can see on this document, line 64 shows us that the PostgreSQL list has an ID input of "VC_1738/VC_1739." This discrepancy was further pointed out by the use of some match functions to see where an ID was missing from either list. This discrepancy accounts for the issue of the missing ID, because of the fact that two IDs were apparently joined with a slash. Below are the two match functions I used in the document:
=MATCH(A2, B$2:B$7665, 0) =MATCH(B2, A$2:A$7665, 0)
.gdb Use in GenMAPP
While the above sections perform quality assurance on the exported Gene Database via verifying ID counts, the "proof in the pudding" is to actually use the Gene Database in GenMAPP. You can follow the instructions in Part 2 of the Vibrio cholerae Microarray Data Analysis to verify that the Gene Database works in GenMAPP. In this case, the emphasis is not on the findings of the data analysis itself, but that the Gene Database functions appropriate in GenMAPP.
For assistance with using the GenMAPP program, the GenMAPP Help is very extensive. To access it within GenMAPP, go to the menu item Help > GenMAPP Help and either browse or search for your topic of interest.
Note: n/a
Putting a gene on the MAPP using the GeneFinder window
- In the main GenMAPP Drafting Board window, left-click on the icon for "Gene" in the upper left corner of the window. Click on the Drafting Board to place the Gene on the MAPP. Now, right-click on the gene to access the GeneFinder window. Type or paste a gene ID into the Gene ID field. Select the appropriate Gene ID system from the drop-down menu and click the Search button. For example, for Vibrio cholerae, you could search for the ID "VC0028", which is an OrderedLocusNames ID. Once the ID has been found, click the OK button to return to the Drafting Board window.
- For the Final Project, you will need to try a sample ID from each of the gene ID systems, not just OrderedLocusNames.
- Open the Backpage by left-clicking on the gene box on the Drafting Board to see if all of the cross-referenced IDs that are supposed to be there are there.
Note: I tried out the search for a gene ID and was able to bring up the Backpage for that ID. The cross-referenced IDs that were supposed to show up were indeed on the page.
Creating an Expression Dataset in the Expression Dataset Manager
- How many of the IDs were imported out of the total IDs in the microarray dataset? How many exceptions were there? Look in the EX.txt file and look at the error codes for the records that were not imported into the Expression Dataset. Do these represent IDs that were present in the UniProt XML, but were somehow not imported? or were they not present in the UniProt XML?
Note: The Expression Dataset Manager reported that there were 121 errors during the conversion. From looking over the error codes, I found that all errors were of the form: Gene not found in OrderedLocusNames or any related system. It seems that these IDs were in fact not present in the UniProt XML.
Coloring a MAPP with expression data
Note: I was able to successfully color the MAPP by coloring the increased and decreased Log Fold Changes.
Running MAPPFinder
Note: After the results had been calculated, a Gene Ontology browser opened showing my results. All of the Gene Ontology terms that have at least 3 genes measured and a p value of less than 0.05 were highlighted yellow. A term with a p value less than 0.05 is considered a "significant" result. Browsed through the tree to see the results.
Documents produced from this run-through can be found here: week 9 docs
Individual Journal Entries
- Week 2
- Week 3
- Week 4
- Week 5
- Week 6
- Week 7
- Week 8
- Week 9
- Week 10
- Week 11
- Week 12
- Week 13
- Week 14
- Week 15