Difference between revisions of "Kevin Wyllie Week 9"
(Pasted in protocol (unedited)) |
(Fixed an answer.) |
||
| (66 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | == Pre-requisites == | ||
| + | |||
| + | This protocol assumes that you are working in a Windows environment. While it is possible to run GenMAPP Builder under the Mac or Linux OS, the end product, a GenMAPP-compatible Gene Database (.gdb), can only be used with the GenMAPP program, which can only be run on Windows. [[Software_Configuration | This set of software has already been installed on the computers in the Seaver 120 computer lab.]] If you want to perform this procedure on your own machine, you must set up your working environment with: | ||
| + | |||
| + | # Any tool that can unpack .gz and .zip files | ||
| + | #* We use [http://www.7-zip.org/ 7-zip] | ||
| + | #* Note that we have found that the native Windows utility cannot reliably unpack .gz files or .zip files containing .jar files. | ||
| + | # [http://www.postgresql.org PostgreSQL] on Windows (http://www.enterprisedb.com/products-services-training/pgdownload) | ||
| + | #* This tutorial was written using PostgreSQL 9.4.x. | ||
| + | # GenMAPP Builder (https://sourceforge.net/projects/xmlpipedb/files/) | ||
| + | # Java JDK 1.8 64-bit | ||
| + | #* [http://www.oracle.com/technetwork/java/javase/downloads/jdk8-downloads-2133151.html Download page] | ||
| + | #* File to download is: jdk-8u65-windows-x64.exe | ||
| + | # GenMAPP 2 can be downloaded [https://github.com/GenMAPPCS/genmapp here]. The file to download is "GenMAPPv2Setup.exe". | ||
| + | # XMLPipeDB ''match'' utility (https://sourceforge.net/projects/xmlpipedb/files/) for counting IDs in XML files | ||
| + | # Microsoft Access or any other tool that can read .mdb files | ||
| + | |||
| + | == Download and Extract Data Source Files == | ||
| + | |||
| + | * Download UniProt XML, GOA, and GO OBO-XML files. | ||
| + | |||
| + | === UniProt XML === | ||
| + | |||
| + | * The UniProt XML file was found on the [http://www.uniprot.org/taxonomy/complete-proteomes UniProt Complete Proteomes] page. | ||
| + | ** "Bacteria" was selected, under the "Superkingdom" heading (on left), followed by "Reference proteome." | ||
| + | ** After scrolling through results, ''Vibrio cholerae'' serotype O1 (strain ATCC 39315 / El Tor Inaba N16961) was found. | ||
| + | ** [http://www.uniprot.org/uniprot/?query=organism:243277 Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961)] was selected, followed by the "download" button at the top of the next page. This opened a dialogue box: | ||
| + | *** "Download all" was selected. | ||
| + | *** "XML" was selected from the "Format" drop down menu. | ||
| + | *** "Compressed" format was chosen. | ||
| + | |||
| + | |||
| + | === GOA === | ||
| + | |||
| + | * [http://www.ebi.ac.uk/GOA This is the UniProt-GOA home page]. | ||
| + | ** The current UniProt-GOA file was downloaded from the [http://ftp.ebi.ac.uk/pub/databases/GO/goa/ UniProt-GOA ftp site]. | ||
| + | ** In the directory that appeared, [http://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/ "proteomes" directory] was chosen. | ||
| + | *** This paged took several minutes to load. | ||
| + | ** [http://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/46.V_cholerae_ATCC_39315.goa ''Vibro cholerae''] was right-clicked, and "save link as" was selected. | ||
| + | *** Left-clicking on this link prompted the file to open in a browser window. | ||
| + | *** The version information was found in the ftp file directory under the "Last modified" column. | ||
| + | |||
| + | === GO OBO-XML === | ||
| + | |||
| + | * Download the GO OBO-XML formatted file from the [http://geneontology.org/page/download-ontology#Legacy_Downloads Gene Ontology download page]. Click on the link for "obo-xml.gz" under the heading "Legacy Downloads." | ||
| + | ** This file is updated daily. You can get the day/time that the file was created from the file properties after you have unzipped the file. | ||
| + | |||
| + | === Extract the UniProt XML and GO OBO-XML files === | ||
| + | |||
| + | * The UniProt XML and GO OBO-XML files were extracted using [http://www.7-zip.org/ 7-zip]. | ||
| + | |||
==Export Information== | ==Export Information== | ||
| − | Version of GenMAPP Builder: | + | Version of GenMAPP Builder: 5 |
Computer on which export was run: HP Compaq 8300 Elite SFF FC | Computer on which export was run: HP Compaq 8300 Elite SFF FC | ||
| Line 7: | Line 64: | ||
Postgres Database name: pgAdminIII | Postgres Database name: pgAdminIII | ||
| − | UniProt XML filename | + | '''UniProt XML filename: [[Media:KW uniprot-organism-243277.zip | uniprot-organism%3A243277]]''' |
| − | * UniProt XML version | + | * UniProt XML version: 2015_10 |
| − | + | * UniProt XML download link: http://www.uniprot.org/uniprot/?query=organism:243277 | |
| − | * Time taken to import: | + | * Time taken to import: 3.02 minutes |
| − | + | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | '''GO OBO-XML filename: [[Media:KW_go_daily-termdb.zip | go_daily-termdb.obo-xml]]''' | |
| − | + | * GO OBO-XML version (The version information can be found in the file properties after the file downloaded from the: 2015 | |
| − | + | * GO OBO-XML download link: http://geneontology.org/page/download-ontology#Legacy_Downloads | |
| − | * Time taken to import: | + | * Time taken to import: 7.68 minutes |
| − | * | + | * Time taken to process: 4.48 |
| − | Name of .gdb file | + | |
| + | '''GOA filename: [[Media:46.V_cholerae_ATCC_39315.goa.zip | 46.V_cholerae_ATCC_39315.goa]]''' | ||
| + | * GOA version: 2015 | ||
| + | * GOA download link: http://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/46.V_cholerae_ATCC_39315.goa | ||
| + | * Time taken to import: 0.06 | ||
| + | |||
| + | |||
| + | '''Name of .gdb file: [[Media:Vc-Std_20151029_KW2.zip | Vc-Std_20151029_KW2.gdb]]''' | ||
* Time taken to export: | * Time taken to export: | ||
| − | ** Start time: | + | ** Start time: 2:40pm |
| − | ** End time: | + | ** End time: 3:52pm |
| − | + | == Download or Update GenMAPP Builder == | |
| + | |||
| + | * [https://github.com/lmu-bioinformatics/xmlpipedb/releases/download/gmbuilder-3.0.0-build-5/gmbuilder-3.0.0-build-5.zip gmbuilder-3.0.0-build-5.zip] was downloaded from the [https://github.com/lmu-bioinformatics/xmlpipedb/releases XMLPipeDB releases page on GitHub]. | ||
| + | * The GenMAPP Builder folder was extracted using [http://www.7-zip.org/ 7-zip]. | ||
| + | * The folder was moved to a safe storage domain. | ||
| + | |||
| + | == Create New Database in PostgreSQL == | ||
| + | |||
| + | * pgAdminIII was launched. | ||
| + | * PostgreSQL 9.4 (localhost:5432) was selected, from the upper left hand side of the window. | ||
| + | ** The appropriate password "Welcome1" was entered. | ||
| + | * "Databases > New database" was selected, and a name was entered for the database. | ||
| + | ** This name included the species name and the date. | ||
| + | * The new database was selected, on left, and the SQL button was clicked on. | ||
| + | * "Open file" was selected from the toolbar, and the unzipped GenMAPP Builder folder was opened in the directory window. | ||
| + | * "gmbuilder.sql" was chosen from within the "sql" folder. | ||
| + | * The "Execute Query" function was chosen (icon resembles a "play" arrow symbol). | ||
| + | ** Notices occurred in the Messages tab (at bottom) indicating that the query returned successfully. | ||
| + | * The query window was closed. To verify that the process worked, the + sign next was clicked, next to the database, followed by the + sign for "public." A "(167)" was seen under the Tables section. | ||
| + | |||
| + | == Configuring GenMAPP Builder to Connect to your PostgreSQL Database == | ||
| + | |||
| + | * gmbuilder.bat was launched. | ||
| + | * "File > Configure database" was selected. | ||
| + | * Under the Database Connections tab the Database Driver defaults to PostgreSQL. Enter information in the following fields: | ||
| + | * The following information was entered into the fields, found under the Database Connections tab: | ||
| + | ** Host or address: localhost | ||
| + | ** Port number: 5432 | ||
| + | ** '''Database name: VCholera_20151027_gmb3build5_KW''' | ||
| + | ** '''Username: postgres''' | ||
| + | ** '''Password: Welcome1''' | ||
| + | |||
| + | == Importing Data into the PostgreSQL Database == | ||
| + | |||
| + | * "File > Import UniProt XML" was selected. | ||
| + | ** The extracted UniProt XML file was chosen in the directory window. | ||
| + | ** The import took 2.96 min (had to redo this one). | ||
| + | * "File > Import GO OBO-XML" was selected. | ||
| + | ** The extracted GO OBO-SML file was chosen in the directory window. | ||
| + | ** The import took 6.88 min. | ||
| + | * "OK" was chosen next to the prompt asking to process the data. | ||
| + | ** The processing took 4.49 min. | ||
| + | * "File > Import GOA" was selected. | ||
| + | ** The downloaded GOA file was chosen in the directory window. | ||
| + | ** The import took 0.06 min. | ||
| + | |||
| + | == Exporting a GenMAPP Gene Database (.gdb) == | ||
| + | |||
| + | * "File > Export to GenMAPP Gene Database" was chosen. | ||
| + | * A name was entered in the Owner field. | ||
| + | * GenMAPP Builder scanned the PostgreSQL database to see what species are available. The appropriate species (''Vibrio cholera'') was chosen. | ||
| + | * To create the GenMAPP database, the "Save GenMAPP Database File As..." button was clicked. | ||
| + | ** In the Save dialog box that appeared, a location was chosen for the file to saved to, and initials were added to the beginning of the default file name. | ||
| + | * The boxes for exporting all Molecular Function, Cellular Component, and Biological Process Gene Ontology Terms were left checked. | ||
| + | * The export process was begun by clicking the "Next" button. | ||
==TallyEngine== | ==TallyEngine== | ||
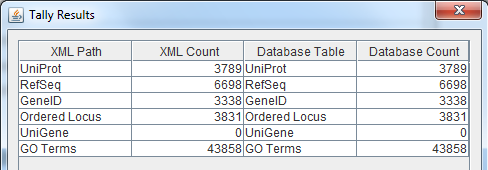
* Run the TallyEngine in GenMAPP Builder and record the number of records for UniProt and GO in the XML data and in the Postgres databases. | * Run the TallyEngine in GenMAPP Builder and record the number of records for UniProt and GO in the XML data and in the Postgres databases. | ||
| − | * | + | * TallyEngine, a tool in GenMAPP Builder, was run to record the number of records for UniProt and GO in the XML data and the Postgres databases. |
| − | + | ** "Tallies > Run XML and Database Tallies for UniProt and GO" was selected. | |
** For more information, [[How_Do_I_Count_Thee%3F_Let_Me_Count_The_Ways | see this page.]] | ** For more information, [[How_Do_I_Count_Thee%3F_Let_Me_Count_The_Ways | see this page.]] | ||
| + | [[file:KW_wk9_screenshot1.png]] | ||
== Using XMLPipeDB match to Validate the XML Results from the TallyEngine== | == Using XMLPipeDB match to Validate the XML Results from the TallyEngine== | ||
| − | [[How_Do_I_Count_Thee%3F_Let_Me_Count_The_Ways | | + | [[How_Do_I_Count_Thee%3F_Let_Me_Count_The_Ways | Instructions found on this page were followed to run XMLPipeDB match.]] |
| − | Are your results the same as you got for the TallyEngine? Why or why not? | + | *'''Are your results the same as you got for the TallyEngine? Why or why not?''' |
| + | ** Using the command: <code>java - jar xmlpipedb-match-1.1.1.jar "VC_A?[0-9][0-9][0-9][0-9]" < uniprot-organism%3A243277.xml</code>, the command line returns <code>Total unique matches: 3831</code>. This number of Gene ID's is the same as for the Tally Engine result. This suggests that the import process was successful. | ||
== Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine== | == Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine== | ||
| Line 50: | Line 163: | ||
For more information, [[How_Do_I_Count_Thee%3F_Let_Me_Count_The_Ways | see this page.]] | For more information, [[How_Do_I_Count_Thee%3F_Let_Me_Count_The_Ways | see this page.]] | ||
| − | + | * Using a variation of a "select count(*) query", counts were conducted in SQL. | |
| + | ** The ''gene/name'' tags in the XML file land in the ''genenametype'' table. A query on this table counting values from this table that were marked as ''ordered locus'' in the XML file matching the pattern ''VC_[0-9][0-9][0-9][0-9]'' would look like this: <code>select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_[0-9][0-9][0-9][0-9]';</code> | ||
| + | * Queries were issued by clicking on the SQL icon in the toolbar, entering the query into the "SQL Editor," and clicking the "Play" button. | ||
| − | |||
| − | + | *'''Are your results the same as reported by the TallyEngine? Why or why not?''' | |
| − | + | ** The above command returns 2737 gene ID's, because it does not account for ID's "VC_A####" | |
| − | [[ | + | ** However, using the command <code>select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_A?[0-9][0-9][0-9][0-9]'</code> returns 3831 gene ID's, which is the same as for the XMLPipeDB Match and Tally Engine Results. This is because adding <code>A?</code> to the command allows pgAdminIII to include gene ID's that either ''do'' or ''don't'' include an "A" following the underscore. |
| − | + | ||
| − | + | ||
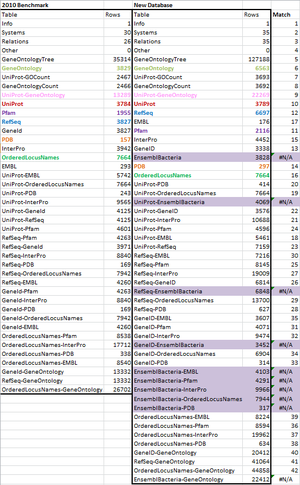
==OriginalRowCounts Comparison== | ==OriginalRowCounts Comparison== | ||
| − | Within the .gdb file, | + | Within the .gdb file, the OriginalRowCounts tables were examined to see if the database had the expected tables with the expected number of records. The 2010 benchmark file and the newly created database are compared below. |
| − | Benchmark .gdb file: | + | '''Benchmark .gdb file:''' [http://sourceforge.net/projects/xmlpipedb/files/V.%20cholerae%20Gene%20Database/V.%20cholerae%2020101022/Vc-Std_External_20101022.zip/download Vc-Std_External_20101022.gdb] |
| − | + | '''OriginalRowCounts table:''' [[media:KW_original_row_counts_from_gdbfile.xlsx]] | |
| + | [[file:Kw_Rowcounts_screenshot_wk9.png|thumb|center]] | ||
| − | + | The OrderedLocusNames table has the same amount of ID's (7664), however plenty of the values are very different, such as GeneOntology, which more than doubled between the benchmark and the new database. Additionally, the new TotalRowCounts table actually has more rows itself. A match function to search for rows that are in the new database but not the old one shows that all "EnsemblBacteria" data is exclusive to the new database. | |
==Visual Inspection== | ==Visual Inspection== | ||
| − | + | A visual inspection of individual tables was done to look for errors. | |
| − | + | ||
| − | + | ||
| − | + | ||
| + | * '''Look at the Systems table. Is there a date in the Date field for all gene ID systems present in the database?''' | ||
| + | ** No. 21 dates are missing. | ||
| + | * '''Open the UniProt, RefSeq, and OrderedLocusNames tables. Scroll down through the table. Do all of the IDs look like they take the correct form for that type of ID?''' | ||
| + | ** UniProt - Yes. All appear to be of uniform format. | ||
| + | ** OrderedLocusNames - No, some have underscores after "VA" and some do not. | ||
| + | ** RefSeq - Yes, assuming different amounts of number characters is not considered to be a deviation from format. | ||
Note: | Note: | ||
==.gdb Use in GenMAPP== | ==.gdb Use in GenMAPP== | ||
| − | |||
| − | |||
===Putting a gene on the MAPP using the GeneFinder window=== | ===Putting a gene on the MAPP using the GeneFinder window=== | ||
| − | + | [[file:Kw_Genefinder_screenshot_wk9.png|center|thumb]] | |
| + | [[file:Kw_Backpage_screenshot_wk9.png|center|thumb]] | ||
| − | + | This seems to work fine. All of the other ID's are present on the backpage. | |
===Creating an Expression Dataset in the Expression Dataset Manager=== | ===Creating an Expression Dataset in the Expression Dataset Manager=== | ||
| − | * How many of the IDs were imported out of the total IDs in the microarray dataset? How many exceptions were there? Look in the EX.txt file and look at the error codes for the records that were not imported into the Expression Dataset. Do these represent IDs that were present in the UniProt XML, but were somehow not imported? | + | * '''How many of the IDs were imported out of the total IDs in the microarray dataset? How many exceptions were there? Look in the EX.txt file and look at the error codes for the records that were not imported into the Expression Dataset. Do these represent IDs that were present in the UniProt XML, but were somehow not imported? Or were they not present in the UniProt XML? |
| − | + | ** I used the Merrell et al. spreadsheet with my newly created .gdb file. Upon creating my expression dataset, I was presented with 121 errors. But this is actually the exact same number of errors reported when the 2010 database was used. This could mean one of two things: either the original GO-OBO-XML, GOA and UniProt files do not contain any significant updates (unlikely given a five-year gap, I think) or there is an error somewhere in the data processing flow.''' | |
| − | + | ** Upon opening the exceptions file, it can be seen that 5221 gene ID's were imported. However, this number includes the 121 errors. | |
| + | ** Adding a filter to column Y ("Errors") and opening the filter menu shows that the only error is "Gene not found in OrderedLocusNames or any related system." Further, applying a text filter for this error message returns 121 rows, verifying that this error message applies to all of the errors. | ||
| + | ** It's hard to believe that the UniProt XML file wouldn't see a ''single'' update over a five year period, especially when there were many just between 2009 and 2010. So I'm inclined to believe this is an import issue. | ||
===Coloring a MAPP with expression data=== | ===Coloring a MAPP with expression data=== | ||
| − | + | [[file:kw_Coloring1_wk9.png|center|thumb]] | |
| + | [[file:Kw_Coloring2_wk9.png|center|thumb]] | ||
| + | |||
| + | This seems to work just fine. | ||
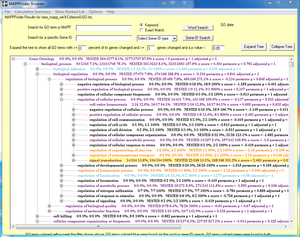
===Running MAPPFinder=== | ===Running MAPPFinder=== | ||
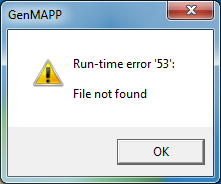
| − | + | [[image:kw_Mappfinder_screenshot.png|thumb|center]] | |
| − | + | [[image:Kw_Error_message_screenshot.png|thumb|center]] | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| + | The MappFinder browser does generate the GO hierarchy, but clicking on ''any'' of the GO terms results in a run-time error. I'm not sure why. | ||
==Links== | ==Links== | ||
Latest revision as of 06:00, 4 November 2015
Contents
- 1 Pre-requisites
- 2 Download and Extract Data Source Files
- 3 Export Information
- 4 Download or Update GenMAPP Builder
- 5 Create New Database in PostgreSQL
- 6 Configuring GenMAPP Builder to Connect to your PostgreSQL Database
- 7 Importing Data into the PostgreSQL Database
- 8 Exporting a GenMAPP Gene Database (.gdb)
- 9 TallyEngine
- 10 Using XMLPipeDB match to Validate the XML Results from the TallyEngine
- 11 Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine
- 12 OriginalRowCounts Comparison
- 13 Visual Inspection
- 14 .gdb Use in GenMAPP
- 15 Links
Pre-requisites
This protocol assumes that you are working in a Windows environment. While it is possible to run GenMAPP Builder under the Mac or Linux OS, the end product, a GenMAPP-compatible Gene Database (.gdb), can only be used with the GenMAPP program, which can only be run on Windows. This set of software has already been installed on the computers in the Seaver 120 computer lab. If you want to perform this procedure on your own machine, you must set up your working environment with:
- Any tool that can unpack .gz and .zip files
- We use 7-zip
- Note that we have found that the native Windows utility cannot reliably unpack .gz files or .zip files containing .jar files.
- PostgreSQL on Windows (http://www.enterprisedb.com/products-services-training/pgdownload)
- This tutorial was written using PostgreSQL 9.4.x.
- GenMAPP Builder (https://sourceforge.net/projects/xmlpipedb/files/)
- Java JDK 1.8 64-bit
- Download page
- File to download is: jdk-8u65-windows-x64.exe
- GenMAPP 2 can be downloaded here. The file to download is "GenMAPPv2Setup.exe".
- XMLPipeDB match utility (https://sourceforge.net/projects/xmlpipedb/files/) for counting IDs in XML files
- Microsoft Access or any other tool that can read .mdb files
Download and Extract Data Source Files
- Download UniProt XML, GOA, and GO OBO-XML files.
UniProt XML
- The UniProt XML file was found on the UniProt Complete Proteomes page.
- "Bacteria" was selected, under the "Superkingdom" heading (on left), followed by "Reference proteome."
- After scrolling through results, Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961) was found.
- Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961) was selected, followed by the "download" button at the top of the next page. This opened a dialogue box:
- "Download all" was selected.
- "XML" was selected from the "Format" drop down menu.
- "Compressed" format was chosen.
GOA
- This is the UniProt-GOA home page.
- The current UniProt-GOA file was downloaded from the UniProt-GOA ftp site.
- In the directory that appeared, "proteomes" directory was chosen.
- This paged took several minutes to load.
- Vibro cholerae was right-clicked, and "save link as" was selected.
- Left-clicking on this link prompted the file to open in a browser window.
- The version information was found in the ftp file directory under the "Last modified" column.
GO OBO-XML
- Download the GO OBO-XML formatted file from the Gene Ontology download page. Click on the link for "obo-xml.gz" under the heading "Legacy Downloads."
- This file is updated daily. You can get the day/time that the file was created from the file properties after you have unzipped the file.
Extract the UniProt XML and GO OBO-XML files
- The UniProt XML and GO OBO-XML files were extracted using 7-zip.
Export Information
Version of GenMAPP Builder: 5
Computer on which export was run: HP Compaq 8300 Elite SFF FC
Postgres Database name: pgAdminIII
UniProt XML filename: uniprot-organism%3A243277
- UniProt XML version: 2015_10
- UniProt XML download link: http://www.uniprot.org/uniprot/?query=organism:243277
- Time taken to import: 3.02 minutes
GO OBO-XML filename: go_daily-termdb.obo-xml
- GO OBO-XML version (The version information can be found in the file properties after the file downloaded from the: 2015
- GO OBO-XML download link: http://geneontology.org/page/download-ontology#Legacy_Downloads
- Time taken to import: 7.68 minutes
- Time taken to process: 4.48
GOA filename: 46.V_cholerae_ATCC_39315.goa
- GOA version: 2015
- GOA download link: http://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/46.V_cholerae_ATCC_39315.goa
- Time taken to import: 0.06
Name of .gdb file: Vc-Std_20151029_KW2.gdb
- Time taken to export:
- Start time: 2:40pm
- End time: 3:52pm
Download or Update GenMAPP Builder
- gmbuilder-3.0.0-build-5.zip was downloaded from the XMLPipeDB releases page on GitHub.
- The GenMAPP Builder folder was extracted using 7-zip.
- The folder was moved to a safe storage domain.
Create New Database in PostgreSQL
- pgAdminIII was launched.
- PostgreSQL 9.4 (localhost:5432) was selected, from the upper left hand side of the window.
- The appropriate password "Welcome1" was entered.
- "Databases > New database" was selected, and a name was entered for the database.
- This name included the species name and the date.
- The new database was selected, on left, and the SQL button was clicked on.
- "Open file" was selected from the toolbar, and the unzipped GenMAPP Builder folder was opened in the directory window.
- "gmbuilder.sql" was chosen from within the "sql" folder.
- The "Execute Query" function was chosen (icon resembles a "play" arrow symbol).
- Notices occurred in the Messages tab (at bottom) indicating that the query returned successfully.
- The query window was closed. To verify that the process worked, the + sign next was clicked, next to the database, followed by the + sign for "public." A "(167)" was seen under the Tables section.
Configuring GenMAPP Builder to Connect to your PostgreSQL Database
- gmbuilder.bat was launched.
- "File > Configure database" was selected.
- Under the Database Connections tab the Database Driver defaults to PostgreSQL. Enter information in the following fields:
- The following information was entered into the fields, found under the Database Connections tab:
- Host or address: localhost
- Port number: 5432
- Database name: VCholera_20151027_gmb3build5_KW
- Username: postgres
- Password: Welcome1
Importing Data into the PostgreSQL Database
- "File > Import UniProt XML" was selected.
- The extracted UniProt XML file was chosen in the directory window.
- The import took 2.96 min (had to redo this one).
- "File > Import GO OBO-XML" was selected.
- The extracted GO OBO-SML file was chosen in the directory window.
- The import took 6.88 min.
- "OK" was chosen next to the prompt asking to process the data.
- The processing took 4.49 min.
- "File > Import GOA" was selected.
- The downloaded GOA file was chosen in the directory window.
- The import took 0.06 min.
Exporting a GenMAPP Gene Database (.gdb)
- "File > Export to GenMAPP Gene Database" was chosen.
- A name was entered in the Owner field.
- GenMAPP Builder scanned the PostgreSQL database to see what species are available. The appropriate species (Vibrio cholera) was chosen.
- To create the GenMAPP database, the "Save GenMAPP Database File As..." button was clicked.
- In the Save dialog box that appeared, a location was chosen for the file to saved to, and initials were added to the beginning of the default file name.
- The boxes for exporting all Molecular Function, Cellular Component, and Biological Process Gene Ontology Terms were left checked.
- The export process was begun by clicking the "Next" button.
TallyEngine
- Run the TallyEngine in GenMAPP Builder and record the number of records for UniProt and GO in the XML data and in the Postgres databases.
- TallyEngine, a tool in GenMAPP Builder, was run to record the number of records for UniProt and GO in the XML data and the Postgres databases.
- "Tallies > Run XML and Database Tallies for UniProt and GO" was selected.
- For more information, see this page.
Using XMLPipeDB match to Validate the XML Results from the TallyEngine
Instructions found on this page were followed to run XMLPipeDB match.
- Are your results the same as you got for the TallyEngine? Why or why not?
- Using the command:
java - jar xmlpipedb-match-1.1.1.jar "VC_A?[0-9][0-9][0-9][0-9]" < uniprot-organism%3A243277.xml, the command line returnsTotal unique matches: 3831. This number of Gene ID's is the same as for the Tally Engine result. This suggests that the import process was successful.
- Using the command:
Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine
For more information, see this page.
- Using a variation of a "select count(*) query", counts were conducted in SQL.
- The gene/name tags in the XML file land in the genenametype table. A query on this table counting values from this table that were marked as ordered locus in the XML file matching the pattern VC_[0-9][0-9][0-9][0-9] would look like this:
select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_[0-9][0-9][0-9][0-9]';
- The gene/name tags in the XML file land in the genenametype table. A query on this table counting values from this table that were marked as ordered locus in the XML file matching the pattern VC_[0-9][0-9][0-9][0-9] would look like this:
- Queries were issued by clicking on the SQL icon in the toolbar, entering the query into the "SQL Editor," and clicking the "Play" button.
- Are your results the same as reported by the TallyEngine? Why or why not?
- The above command returns 2737 gene ID's, because it does not account for ID's "VC_A####"
- However, using the command
select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_A?[0-9][0-9][0-9][0-9]'returns 3831 gene ID's, which is the same as for the XMLPipeDB Match and Tally Engine Results. This is because addingA?to the command allows pgAdminIII to include gene ID's that either do or don't include an "A" following the underscore.
OriginalRowCounts Comparison
Within the .gdb file, the OriginalRowCounts tables were examined to see if the database had the expected tables with the expected number of records. The 2010 benchmark file and the newly created database are compared below.
Benchmark .gdb file: Vc-Std_External_20101022.gdb
OriginalRowCounts table: media:KW_original_row_counts_from_gdbfile.xlsx
The OrderedLocusNames table has the same amount of ID's (7664), however plenty of the values are very different, such as GeneOntology, which more than doubled between the benchmark and the new database. Additionally, the new TotalRowCounts table actually has more rows itself. A match function to search for rows that are in the new database but not the old one shows that all "EnsemblBacteria" data is exclusive to the new database.
Visual Inspection
A visual inspection of individual tables was done to look for errors.
- Look at the Systems table. Is there a date in the Date field for all gene ID systems present in the database?
- No. 21 dates are missing.
- Open the UniProt, RefSeq, and OrderedLocusNames tables. Scroll down through the table. Do all of the IDs look like they take the correct form for that type of ID?
- UniProt - Yes. All appear to be of uniform format.
- OrderedLocusNames - No, some have underscores after "VA" and some do not.
- RefSeq - Yes, assuming different amounts of number characters is not considered to be a deviation from format.
Note:
.gdb Use in GenMAPP
Putting a gene on the MAPP using the GeneFinder window
This seems to work fine. All of the other ID's are present on the backpage.
Creating an Expression Dataset in the Expression Dataset Manager
- How many of the IDs were imported out of the total IDs in the microarray dataset? How many exceptions were there? Look in the EX.txt file and look at the error codes for the records that were not imported into the Expression Dataset. Do these represent IDs that were present in the UniProt XML, but were somehow not imported? Or were they not present in the UniProt XML?
- I used the Merrell et al. spreadsheet with my newly created .gdb file. Upon creating my expression dataset, I was presented with 121 errors. But this is actually the exact same number of errors reported when the 2010 database was used. This could mean one of two things: either the original GO-OBO-XML, GOA and UniProt files do not contain any significant updates (unlikely given a five-year gap, I think) or there is an error somewhere in the data processing flow.
- Upon opening the exceptions file, it can be seen that 5221 gene ID's were imported. However, this number includes the 121 errors.
- Adding a filter to column Y ("Errors") and opening the filter menu shows that the only error is "Gene not found in OrderedLocusNames or any related system." Further, applying a text filter for this error message returns 121 rows, verifying that this error message applies to all of the errors.
- It's hard to believe that the UniProt XML file wouldn't see a single update over a five year period, especially when there were many just between 2009 and 2010. So I'm inclined to believe this is an import issue.
Coloring a MAPP with expression data
This seems to work just fine.
Running MAPPFinder
The MappFinder browser does generate the GO hierarchy, but clicking on any of the GO terms results in a run-time error. I'm not sure why.
Links
- Kevin Wyllie Week 2 (See the original assignment and class journal.)
- Kevin Wyllie Week 3 (See the original assignment and class journal.)
- Kevin Wyllie Week 4 (See the original assignment and class journal.)
- Kevin Wyllie Week 5 (See the original assignment and class journal.)
- Kevin Wyllie Week 6 (See the original assignment and class journal.)
- Kevin Wyllie Week 7 (See the original assignment and class journal.)
- Kevin Wyllie Week 8 (See the original assignment and class journal.)
- Kevin Wyllie Week 9 (See the original assignment and class journal.)
- Kevin Wyllie Week 10 (See the original assignment.)
- Kevin Wyllie Week 11 (See the original assignment.)
- Kevin Wyllie Week 12 (See the original assignment.)
- Kevin Wyllie Week 14 (See the original assignment.)
- Kevin Wyllie Week 15 (See the original assignment.)