|
|
| Line 1: |
Line 1: |
| − | ==Initial Preparations==
| |
| − | ''List of required programs was initially sourced from the [[Running GenMAPP Builder | Running GenMAPP Builder page]]''
| |
| − |
| |
| − | ''One of the final goals is to perform an export of a ''Vibrio cholerae GenMAPP Gene Database''
| |
| − |
| |
| − | In preparation for this assignment, it was ensured that these programs were installed on a Windows workstation:
| |
| − | *[http://www.7-zip.org/ 7-zip] for the unpacking of compressed files
| |
| − | *[http://www.enterprisedb.com/products-services-training/pgdownload PostgreSQL]
| |
| − | *[https://sourceforge.net/projects/xmlpipedb/files/ GenMAPP Builder]
| |
| − | *[http://www.oracle.com/technetwork/java/javase/downloads/jdk8-downloads-2133151.html Java JDK 1.8 64-bit] (download jdk-8u65-windows-x64.exe)
| |
| − | *[https://github.com/GenMAPPCS/genmapp GenMAPP 2] (download GenMAPPv2Setup.exe)
| |
| − | *[https://sourceforge.net/projects/xmlpipedb/files/) XMLPipeDB ''match'' utility]
| |
| − | *A program that is able to read .mdb files (such as Microsoft Access)
| |
| − | ==Downloading the Required Files==
| |
| − | ''List of required files was initially sourced from the [[Running GenMAPP Builder | Running GenMAPP Builder page]]''
| |
| − | ===Retrieving the UniProt XML file, Performed on 10/27===
| |
| − | *The [http://www.uniprot.org/taxonomy/complete-proteomes UniProt Complete Proteomes] page was entered
| |
| − | *The Superkingdom ''Bacteria'' was selected and ''Reference proteome'' was clicked on (72 results currently)
| |
| − | *"vibrio cholerae" was added to the search bar and search was clicked upon. The results were filtered to the only proteome of interest (''Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961)'')
| |
| − | *The result was clicked upon and, on the result page, ''UniProtKB'' was clicked upon in the "Map to" section (on left of the page)
| |
| − | *On the ''UniProtKB results'' page, Download was clicked; in the box that appeared, download all was selected, the format was set to XML, and the file was set to be compressed.
| |
| − | ===Retrieving the GOA file, Performed on 10/27===
| |
| − | *The [http://ftp.ebi.ac.uk/pub/databases/GO/goa/ UniProt-GOA ftp site] was entered
| |
| − | *The link to the "proteomes" directory was clicked in the main directory
| |
| − | *In "proteomes", the file V_cholerae_ATCC_39315.goa was searched for, found, and downloaded by using right-click with "Save link as"
| |
| − | ===Retrieving the GO OBO-XML file, Performed on 10/27===
| |
| − | *The GO OBO-XML file was downloaded from the [http://geneontology.org/page/download-ontology#Legacy_Downloads Gene Ontology download page]
| |
| − | *"obo-xml.gz" was selected/clicked under Legacy Downloads
| |
| − | ===Downloading/Updating GenMAPP Builder, Performed on 10/27===
| |
| − | *The files were downloaded from the [https://github.com/lmu-bioinformatics/xmlpipedb/releases XMLPipeDB releases page on GitHub]
| |
| − | *[https://github.com/lmu-bioinformatics/xmlpipedb/releases/download/gmbuilder-3.0.0-build-5/gmbuilder-3.0.0-build-5.zip Download link for gmbuilder-3.0.0-build-5 (the version used in this assignment)]
| |
| − | **'''All of the downloaded files, if compressed, were extracted using 7-Zip. All required files were placed in one folder'''
| |
| − | *''Downloaded on 10/27, Summary''
| |
| − | **The complete proteome for ''V. cholerae'' was downloaded from [http://www.uniprot.org/uniprot/?query=organism:243277 UniProtKB] in the XML format
| |
| − | **The GOA file for ''V. cholerae'' was downloaded from this [http://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/46.V_cholerae_ATCC_39315.goa link]
| |
| − | **The GO OBO-XML formatted file for ''V. cholerae'' was downloaded from the [http://geneontology.org/page/download-ontology#Legacy_Downloads GO website]
| |
| − | **The most recent version (3.0.0, build 5) of gmBuilder was downloaded from [https://github.com/lmu-bioinformatics/xmlpipedb/releases/download/gmbuilder-3.0.0-build-5/gmbuilder-3.0.0-build-5.zip GitHub]
| |
| − |
| |
| − | ==Export Process==
| |
| − | ===Creating a New Database in PostgreSQL===
| |
| − | *Steps taken were sourced from the [[Running GenMAPP Builder | Running GenMAPP Builder page]]
| |
| − | *pgAdmin III was launched and a connection to the server was made. "Databases" was right clicked and select "New Database..." was chosen. The database was given a name, V.cholerae_20151027_gmb3build5, and OK was clicked.
| |
| − | *The new database was selected and the Query Tool was launched. Open File was clicked in the Query Tool and ''gmbuilder.sql'' in the ''gmbuilder-3.0.0-build-5'' folder (within the ''sql'' folder) was selected. Upon selection of that file, a query was loaded into Query Tool and it was subsequently executed by clicking the green "Execute Query" arrow
| |
| − | *This query populates the created database with all of its tables. In order to ensure that the query properly worked, it was checked that 167 tables existed in the database
| |
| − | ===Importing Data===
| |
| − | *gmbuilder.bat in the gmbuilder folder was launched
| |
| − | *Under file -> configure database, the host was left as localhost, the port number was left as 5432, database name was set to ''V.cholerae_20151027_gmb3build5'', Username was set to BL, Password was set to the password of the PostgreSQL database that was recently created. OK was clicked.
| |
| − | ===Data Import into ''V.cholerae_20151027_gmb3build5''===
| |
| − | *File -> Import UniProt XML was selected
| |
| − | **The UniProt XML file that was previously extracted was chosen, open was clicked. The import process was allowed to proceed uninterrupted.
| |
| − | *File -> Import GO OBO-XML was selected
| |
| − | **The GO OBO-XML that was previously extracted was chosen, open was clicked. The import process was allowed to proceed uninterrupted.
| |
| − | *File -> Import GOA was selected
| |
| − | **The GOA file that was downloaded previously was chosen, open was clicked, and the import process was allowed to proceed uninterrupted.
| |
| − | ===Exporting a GenMAPP Gene Database (.gdb file)===
| |
| − | *File -> Export to GenMAPP Gene Database was selected
| |
| − | *BL was typed into the Owner field. The species of interest was selected for export (''V. cholerae'')
| |
| − | *Next was clicked, the create GenMAPP database file/location was selected, and the boxes for the exporting of Molecular Function, Cellular Component, and Biological Process Gene Ontology Terms were left checked. The export process was initialized by clicking next; the windows were left open for the program to continue and finish with the export process (was estimated to take somewhere between 1-2 hrs).
| |
| − | ==Gene Database Testing Report==
| |
| − | ===Export Information===
| |
| − | Version of GenMAPP Builder: 3.0.0-build-5
| |
| − |
| |
| − | Computer on which export was run: Workstation in Seaver 120
| |
| − |
| |
| − | Postgres Database name: V.cholerae_20151027_gmb3build5
| |
| − |
| |
| − | UniProt XML filename: [[Media:uniprot-organism_BL_20151027.xml.gz|compressed uniprot-organism%3A243277_BL_20151027.xml]]
| |
| − | * UniProt XML version (found out using the [http://www.uniprot.org/help/?fil=section:news UniProt News Page]: UniProt release 2015_10
| |
| − | * UniProt XML download link: http://www.uniprot.org/uniprot/?query=organism:243277#
| |
| − | * Time taken to import: 2.99 minutes
| |
| − | ** Note: Import times were as expected; nothing unusual occurred during the import process.
| |
| − |
| |
| − | GO OBO-XML filename: [[Media:BL 20151027 go daily-termdb.zip|compressed BL_20151027_go_daily-termdb.obo-xml]]
| |
| − | * GO OBO-XML version (derived from the date modified on the file, itself): Date modified - ''10/27/2015 2:24 AM''
| |
| − | * GO OBO-XML download link: [http://geneontology.org/page/download-ontology#Legacy_Downloads GO website]
| |
| − | * Time taken to import: 6.98 minutes
| |
| − | * Time taken to process: 4.54 minutes
| |
| − | ** Note: Import times were as expected; nothing unusual occurred during the import process.
| |
| − |
| |
| − | GOA filename: [[Media:46.V_cholerae_ATCC_39315_BL_20151027.zip| compressed 46.V_cholerae_ATCC_39315_BL_20151027.goa]]
| |
| − | * GOA version (taken from [http://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/ here]: Last Update - ''13-Oct-2015 07:31''
| |
| − | * GOA download link: [http://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/46.V_cholerae_ATCC_39315.goa link]
| |
| − | * Time taken to import: 0.06 minutes
| |
| − | ** Note: Import times were as expected; nothing unusual occurred during the import process.
| |
| − |
| |
| − | Name of .gdb file: [[Media:Vc-Std_20151027_BL.zip | compressed Vc-Std_20151027_BL.gdb]]
| |
| − | * Time taken to export: one hour and nineteen minutes
| |
| − | ** Start time: 3:56
| |
| − | ** End time: 5:15
| |
| − | Note: Export times were as expected; nothing unusual occurred during the export process. Program windows, however, were closed by the time the export product was observed; the time taken to export was found out by observing the time on the Date Modified section of the produced file. The found end time was in line with the times found by others (ranged around an hour and 15 minutes)
| |
| − |
| |
| − | ===Using TallyEngine to test the newly created database===
| |
| − | *PostgreSQL was initialized through pgAdmin III and the database V.cholerae_20151027_gmb3build5 was left running
| |
| − | *GenMAPP builder was booted and ''Run XML and Database Tallies for UniProt and GO'' was selected under the ''Tallies'' menu item; the UniProt XML and GO files that were imported were chosen
| |
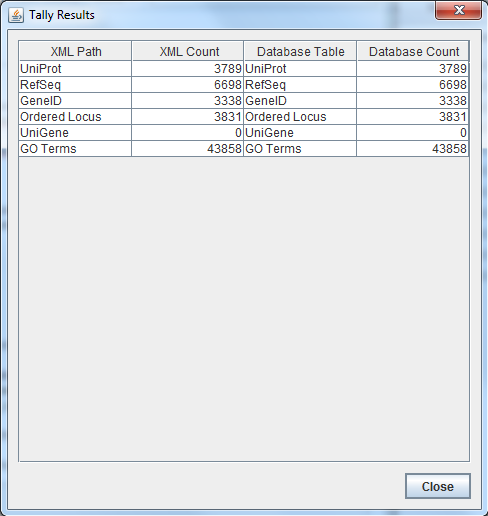
| − | *'''Results of TallyEngine:'''
| |
| − | *[[File:TallyEngine_Results_BL_20151029.png]]
| |
| − |
| |
| − | ===Using XMLPipeDB match to Validate the XML Results from TallyEngine===
| |
| − | *The Windows command line was launched (cmd.exe)
| |
| − | *This set of commands was inputted into the command line in order to utilize XMLPipeDB match:
| |
| − | <code>java -jar xmlpipedb-match-1.1.1.jar "VC_[0-9][0-9][0-9][0-9]" < uniprot-organism%3A243277.xml</code>
| |
| − | **NOTE: Prior to executing the command, the folder that held the files and xmlpipedb-match-1.1.1.jar was entered through the Windows command line (a set of CD commands was used in order to enter the correct directory)
| |
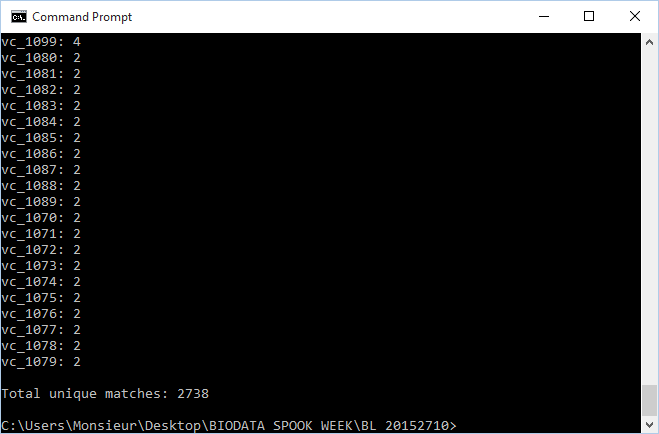
| − | *2738 unique matches were found through XMLPipeDB match
| |
| − | **[[File:BL MATCHUTILITY WEEK9.png]]
| |
| − | Are your results the same as you got for the TallyEngine? Why or why not?
| |
| − | *These results are not the same as what was found using TallyEngine (3831 results were found through TallyEngine). It is suspected that the match command pattern is not fully representative of everything that falls under ''OrderedLocusNames'' (the fault, it is believed, lies with match and not with TallyEngine)
| |
| − |
| |
| − | ===Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine===
| |
| − | *''pgAdmin III'' was booted and all of the necessary connections were made
| |
| − | *It was realized that the ''gene/name'' tags in the XML file end up in the ''genenametype'' table (source: [[How_Do_I_Count_Thee%3F_Let_Me_Count_The_Ways |the wiki page regarding database quality analysis]]
| |
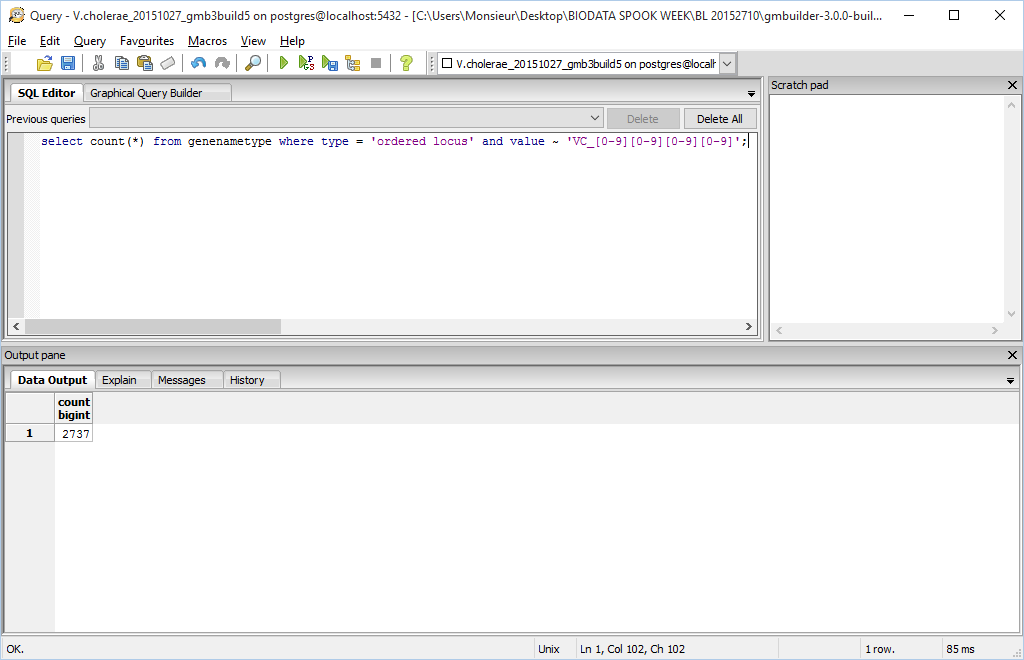
| − | *In ''pgAdmin III'', the query <code>select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_[0-9][0-9][0-9][0-9]';</code> was issued via the SQL Query menu in order to further validate the PostgreSQL Database Results according to TallyEngine
| |
| − | *2737 unique matches were found in ''pgAdmin III'' (postgres database results). These results are not the same as what was found by TallyEngine and are slightly different from what was found using XMLPipeDB match (1 off). It seems that the results are really close to what the Match utility found because it utilizes the same pattern ('VC_[0-9][0-9][0-9][0-9]')
| |
| − | **[[File:BL PSQL COUNT WEEK9.png]]
| |
| − |
| |
| − | ===OriginalRowCounts Comparison using Microsoft Access===
| |
| − | *The exported ''Vc-Std_20151027_BL.gdb'' file was opened using Microsoft Access
| |
| − | *7664 unique matches found using Microsoft Access were found by observing the row count for the ''OrderedLocusNames'' table. This number of unique matches is very different from what was found using the other methods (more than double of what was found using TallyEngine, which is 3831 unique results). It is believed that the ''OrderedLocusNames'' table contains two sets of data (different "versions") with each harboring one value/ID that is unaccounted for by the other count validation methods.
| |
| − |
| |
| − | ===Visual Inspection===
| |
| − | ====Observing the 'OrderedLocusNames' table, and further modifications====
| |
| − | *The database was opened in a program that is able to read .mdb files, such as Microsoft Access
| |
| − | *Looking at the OrderedLocusNames column, it was noticed that some of the entries contain underscores after the ''VC'' and others do not. By filtering so that only the entries with underscores appear, it was noticed that there are 3832 total entries with underscores. 3832 is half of 7664, which is the total amount of entries in the OrderedLocusNames table; this suggests that there exists two different versions of the data (with underscores and without). It was also noticed that both versions of the data (underscores and without) have some entries that include a letter A after the ''VC'' portion; through filtering, it was noticed that the entries with A's account for 1095 of the entries in both sets of data.
| |
| − | *Including the As in the command for the Match utility, <code>java -jar xmlpipedb-match-1.1.1.jar "VC_A?[0-9][0-9][0-9][0-9]" < uniprot-organism%3A243277.xml</code>, resulting in a result of 3831 unique matches; 3831 is an exact match of what was found using TallyEngine.
| |
| − | *Modifying the SQL query so that the pattern includes the entries with A's, by using <code>select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_A?[0-9][0-9][0-9][0-9]';</code>, showed that there are 3831 entries (the number is in line with what TallyEngine found).
| |
| − | *It now appears that there is one entry in both data sets (underscore and non-underscore) that remains unaccounted for
| |
| − | ===Using Microsoft Excel to Compare ID Lists===
| |
| − | *The 'OrderedLocusNames' table was first put in ascending order (lower ID #s come first) and was then exported in a format that Excel can read (was then opened with Excel)
| |
| − | *<code>java -jar xmlpipedb-match-1.1.1.jar "VC_A?[0-9][0-9][0-9][0-9]" < uniprot-organism%3A243277.xml > ordered_locus_match_results_BL.txt</code> was used via the Windows command line in order to export the ID lists with the underscores to a text file (one half of all of the IDs)
| |
| − | *The Match utility exported text file was opened via Excel and the colon ('':'') was set as the delimiter (since a colon separates the match count from the ID)
| |
| − | *pgAdmin III was booted up and the proper database was selected (V.cholerae_20151027_gmb3build5); the SQL query <code>select value from genenametype where type = 'ordered locus';</code> was inputted and "Execute query, write result to file" was clicked. Through these steps, the list of IDs from PostgreSQL was exported in an Excel friendly format (such as .csv)
| |
| − | *The column of IDs from 'OrderedLocusNames', the one exported from the Match utility, and the one that was exported from PostgreSQL, were put side by side in a new Excel document with no spaces between them; it was ensured that each column was in ascending order. The column of IDs from 'OrderedLocusNames' was given the label ' 'IDs FROM 'ORDEREDLOCUSNAMES' ', the one that was exported from the Match utility was given the label ''IDs FROM MATCH'', and the one that was exported from PostgreSQL was given the label ''IDs FROM postgreSQL''
| |
| − | *A third column was inserted directly to the right of the last column with the heading ''MATCH''
| |
| − | *''IDs FROM 'ORDEREDLOCUSNAMES'' was additionally labeled with a number 1, ''IDs FROM MATCH'' was additionally labeled with a number 2, and ''IDs FROM postgreSQL'' was additionally labeled with a number 3
| |
| − | *6 empty adjacent columns were reserved for Excel MATCH commands (which look up ID values from one column in another column/range); MATCH outputs the position where the matched value was found (starting from 1) or '''#N/A''' if there existed no match at that position
| |
| − | *6 empty columns were labeled directly to the right (adjacent) to ''IDs FROM postgreSQL (3)''; in order, from left to right, the column labels typed in were ''MATCH:1 to 2'', ''MATCH:1 to 3'', ''MATCH:2 to 1'', ''MATCH:2 to 3'', ''MATCH:3 to 1'', and ''MATCH:3 to 2''
| |
| − | **NOTE: In the MATCH column labels, the numbers 1, 2, and 3 correspond to distinct ID columns; 1 represents ''IDs FROM 'ORDEREDLOCUSNAMES'', 2 represents ''IDs FROM MATCH'', and 3 represents ''IDs FROM postgreSQL''
| |
| − | *MATCH commands were then written for each of the 6 MATCH columns and applied to the entirety of each MATCH column; the basic format is <code>=MATCH(VALUE TO LOOK-UP, RANGE/COLUMN WHERE THE LOOKING-UP OF A VALUE TAKES PLACE, "MATCH TYPE" [0 in this case])</code>. The purpose of these MATCH commands is to compare the three different ID lists (with each other)
| |
| − |
| |
| − | '''ALL MATCH COMMANDS:'''
| |
| − | Format - '''Column Label''' : <code>MATCH Command</code> (in first "cell")
| |
| − | '''MATCH:1 to 2''' : <code>=MATCH(A2, B$2:B$3832,0)</code>
| |
| − | '''MATCH:1 to 3''' : <code>=MATCH(A2, C$2:C$3832,0)</code>
| |
| − | '''MATCH:2 to 1''' : <code>=MATCH(B2, A$2:A$3833,0)</code>
| |
| − | '''MATCH:2 to 3''' : <code>=MATCH(B2, C$2:C$3832,0)</code>
| |
| − | '''MATCH:3 to 1''' : <code>=MATCH(C2, A$2:A$3833,0)</code>
| |
| − | '''MATCH:3 to 2''' : <code>=MATCH(C2, B$2:B$3832,0)</code>
| |
| − | ====MATCH results analysis====
| |
| − | *''ctrl+F'' was utilized in order to find instances of '''#N/A''' among all of the MATCH columns (some options included, Look In: ''Values'', Find What: ''#N/A'', Find All); eight cases of '''#N/A''' were found
| |
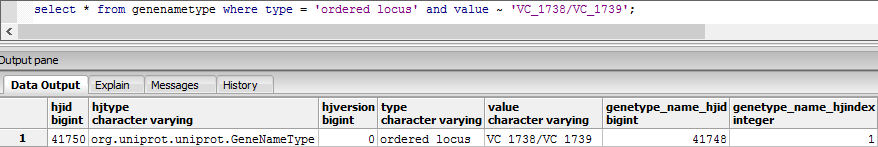
| − | *By observing the cases of '''#N/A''' (and the corresponding IDs), it was realized that 6 of the cases were due to the presence of '''VC_1738/VC_1739''' in position '''1711''' within the ''IDs FROM postgreSQL (3)'' column
| |
| − | *The last two cases of '''#N/A''' were found to be the result of the presence of '''VC_A0360.1''' in position '''3093''' within the ''IDs FROM 'ORDEREDLOCUSNAMES (1)'' column, and in position '''3092''' within the ''IDs FROM postgreSQL (3)'' column
| |
| − | *It seems apparent that there exist some entries in the 'ORDEREDLOCUSNAMES' that are not accounted for by the MATCH utility ('''VC_A0360.1'''); it is also clear that there are two entries acting as one entry within the postgreSQL output ('''VC_1738/VC_1739'''). '''VC_A0360.1''' also exists within the postgreSQL output.
| |
| − | ===Further Quality Assurance Work===
| |
| − | *Using the Match utility, <code>java -jar xmlpipedb-match-1.1.1.jar "VC_A?[0-9][0-9][0-9][0-9](.1)?" < uniprot-organism%3A243277.xml</code> was inputted into the Windows command line in order to get all 3832 possible unique matches (includes '''VC_A0360.1''')
| |
| − | *In pgAdmin III, the SQL query <code>select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_A?[0-9][0-9][0-9][0-9](.1)?';</code> returns the same count as the previously attempted query (the previously attempted query appears to have taken in account to the presence of '''VC_A0360.1'''); the issue that is preventing a count of 3832 seems to be the presence of '''VC_1738/VC_1739''' in one of the rows:
| |
| − | **[[File:Pgsql_issuerow_BL_wk9.png]]
| |
| − | ==.gdb Use in GenMAPP==
| |
| − | *Some of the protocol from [http://www.openwetware.org/wiki/BIOL367/F10:GenMAPP_and_MAPPFinder_Protocols Part 2 of the ''Vibrio cholerae'' Microarray Data Analysis] was used as a reference for this portion of the assignment
| |
| − | *''Vc-Std_20151027_BL.gdb'', the recently created ''Vibrio cholerae'' database, was placed within the Gene Databases folder of the GenMAPP directory (the folder is within the GenMAPP 2 Data folder)
| |
| − | *GenMAPP (Version 2.1) was launched
| |
| − | *The new gene database was loaded by going into ''Data > Choose Gene Database''
| |
| − | *The tab deliminated GenMAPP formatted [[Media:Merrell_Compiled_Raw_Data_Vibrio_BL_20151015.txt|data]] from week 8 was loaded into GenMAPP through ''Data > Expression Dataset Manager > Expression Datasets > New Dataset > Merrell_Compiled_Raw_Data_Vibrio_BL_20151015.txt''
| |
| − |
| |
| − |
| |
| − |
| |
| − | While the above sections perform quality assurance on the exported Gene Database via verifying ID counts, the "proof in the pudding" is to actually use the Gene Database in GenMAPP. You can follow the instructions in [http://www.openwetware.org/wiki/BIOL367/F10:GenMAPP_and_MAPPFinder_Protocols Part 2 of the ''Vibrio cholerae'' Microarray Data Analysis] to verify that the Gene Database works in GenMAPP. In this case, the emphasis is not on the findings of the data analysis itself, but that the Gene Database functions appropriate in GenMAPP.
| |
| − |
| |
| − | Note:
| |
| − |
| |
| − | ===Putting a gene on the MAPP using the GeneFinder window===
| |
| − |
| |
| − | * Try a sample ID from each of the gene ID systems. Open the Backpage and see if all of the cross-referenced IDs that are supposed to be there are there.
| |
| − |
| |
| − | Note:
| |
| − |
| |
| − | ===Creating an Expression Dataset in the Expression Dataset Manager===
| |
| − |
| |
| − | * How many of the IDs were imported out of the total IDs in the microarray dataset? How many exceptions were there? Look in the EX.txt file and look at the error codes for the records that were not imported into the Expression Dataset. Do these represent IDs that were present in the UniProt XML, but were somehow not imported? or were they not present in the UniProt XML?
| |
| − |
| |
| − | Note:
| |
| − |
| |
| − | ===Coloring a MAPP with expression data===
| |
| − |
| |
| − | Note:
| |
| − |
| |
| − | ===Running MAPPFinder===
| |
| − |
| |
| − | Note:
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − | ==Initial Preparations==
| |
| − | ''List of required programs was initially sourced from the [[Running GenMAPP Builder | Running GenMAPP Builder page]]''
| |
| − |
| |
| − | ''One of the final goals is to perform an export of a ''Vibrio cholerae GenMAPP Gene Database''
| |
| − |
| |
| − | In preparation for this assignment, it was ensured that these programs were installed on a Windows workstation:
| |
| − | *[http://www.7-zip.org/ 7-zip] for the unpacking of compressed files
| |
| − | *[http://www.enterprisedb.com/products-services-training/pgdownload PostgreSQL]
| |
| − | *[https://sourceforge.net/projects/xmlpipedb/files/ GenMAPP Builder]
| |
| − | *[http://www.oracle.com/technetwork/java/javase/downloads/jdk8-downloads-2133151.html Java JDK 1.8 64-bit] (download jdk-8u65-windows-x64.exe)
| |
| − | *[https://github.com/GenMAPPCS/genmapp GenMAPP 2] (download GenMAPPv2Setup.exe)
| |
| − | *[https://sourceforge.net/projects/xmlpipedb/files/) XMLPipeDB ''match'' utility]
| |
| − | *A program that is able to read .mdb files (such as Microsoft Access)
| |
| − | ==Downloading the Required Files==
| |
| − | ''List of required files was initially sourced from the [[Running GenMAPP Builder | Running GenMAPP Builder page]]''
| |
| − | ===Retrieving the UniProt XML file, Performed on 10/27===
| |
| − | *The [http://www.uniprot.org/taxonomy/complete-proteomes UniProt Complete Proteomes] page was entered
| |
| − | *The Superkingdom ''Bacteria'' was selected and ''Reference proteome'' was clicked on (72 results currently)
| |
| − | *"vibrio cholerae" was added to the search bar and search was clicked upon. The results were filtered to the only proteome of interest (''Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961)'')
| |
| − | *The result was clicked upon and, on the result page, ''UniProtKB'' was clicked upon in the "Map to" section (on left of the page)
| |
| − | *On the ''UniProtKB results'' page, Download was clicked; in the box that appeared, download all was selected, the format was set to XML, and the file was set to be compressed.
| |
| − | ===Retrieving the GOA file, Performed on 10/27===
| |
| − | *The [http://ftp.ebi.ac.uk/pub/databases/GO/goa/ UniProt-GOA ftp site] was entered
| |
| − | *The link to the "proteomes" directory was clicked in the main directory
| |
| − | *In "proteomes", the file V_cholerae_ATCC_39315.goa was searched for, found, and downloaded by using right-click with "Save link as"
| |
| − | ===Retrieving the GO OBO-XML file, Performed on 10/27===
| |
| − | *The GO OBO-XML file was downloaded from the [http://geneontology.org/page/download-ontology#Legacy_Downloads Gene Ontology download page]
| |
| − | *"obo-xml.gz" was selected/clicked under Legacy Downloads
| |
| − | ===Downloading/Updating GenMAPP Builder, Performed on 10/27===
| |
| − | *The files were downloaded from the [https://github.com/lmu-bioinformatics/xmlpipedb/releases XMLPipeDB releases page on GitHub]
| |
| − | *[https://github.com/lmu-bioinformatics/xmlpipedb/releases/download/gmbuilder-3.0.0-build-5/gmbuilder-3.0.0-build-5.zip Download link for gmbuilder-3.0.0-build-5 (the version used in this assignment)]
| |
| − | **'''All of the downloaded files, if compressed, were extracted using 7-Zip. All required files were placed in one folder'''
| |
| − | *''Downloaded on 10/27, Summary''
| |
| − | **The complete proteome for ''V. cholerae'' was downloaded from [http://www.uniprot.org/uniprot/?query=organism:243277 UniProtKB] in the XML format
| |
| − | **The GOA file for ''V. cholerae'' was downloaded from this [http://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/46.V_cholerae_ATCC_39315.goa link]
| |
| − | **The GO OBO-XML formatted file for ''V. cholerae'' was downloaded from the [http://geneontology.org/page/download-ontology#Legacy_Downloads GO website]
| |
| − | **The most recent version (3.0.0, build 5) of gmBuilder was downloaded from [https://github.com/lmu-bioinformatics/xmlpipedb/releases/download/gmbuilder-3.0.0-build-5/gmbuilder-3.0.0-build-5.zip GitHub]
| |
| − |
| |
| − | ==Export Process==
| |
| − | ===Creating a New Database in PostgreSQL===
| |
| − | *Steps taken were sourced from the [[Running GenMAPP Builder | Running GenMAPP Builder page]]
| |
| − | *pgAdmin III was launched and a connection to the server was made. "Databases" was right clicked and select "New Database..." was chosen. The database was given a name, V.cholerae_20151027_gmb3build5, and OK was clicked.
| |
| − | *The new database was selected and the Query Tool was launched. Open File was clicked in the Query Tool and ''gmbuilder.sql'' in the ''gmbuilder-3.0.0-build-5'' folder (within the ''sql'' folder) was selected. Upon selection of that file, a query was loaded into Query Tool and it was subsequently executed by clicking the green "Execute Query" arrow
| |
| − | *This query populates the created database with all of its tables. In order to ensure that the query properly worked, it was checked that 167 tables existed in the database
| |
| − | ===Importing Data===
| |
| − | *gmbuilder.bat in the gmbuilder folder was launched
| |
| − | *Under file -> configure database, the host was left as localhost, the port number was left as 5432, database name was set to ''V.cholerae_20151027_gmb3build5'', Username was set to BL, Password was set to the password of the PostgreSQL database that was recently created. OK was clicked.
| |
| − | ===Data Import into ''V.cholerae_20151027_gmb3build5''===
| |
| − | *File -> Import UniProt XML was selected
| |
| − | **The UniProt XML file that was previously extracted was chosen, open was clicked. The import process was allowed to proceed uninterrupted.
| |
| − | *File -> Import GO OBO-XML was selected
| |
| − | **The GO OBO-XML that was previously extracted was chosen, open was clicked. The import process was allowed to proceed uninterrupted.
| |
| − | *File -> Import GOA was selected
| |
| − | **The GOA file that was downloaded previously was chosen, open was clicked, and the import process was allowed to proceed uninterrupted.
| |
| − | ===Exporting a GenMAPP Gene Database (.gdb file)===
| |
| − | *File -> Export to GenMAPP Gene Database was selected
| |
| − | *BL was typed into the Owner field. The species of interest was selected for export (''V. cholerae'')
| |
| − | *Next was clicked, the create GenMAPP database file/location was selected, and the boxes for the exporting of Molecular Function, Cellular Component, and Biological Process Gene Ontology Terms were left checked. The export process was initialized by clicking next; the windows were left open for the program to continue and finish with the export process (was estimated to take somewhere between 1-2 hrs).
| |
| − | ==Gene Database Testing Report==
| |
| − | ===Export Information===
| |
| − | Version of GenMAPP Builder: 3.0.0-build-5
| |
| − |
| |
| − | Computer on which export was run: Workstation in Seaver 120
| |
| − |
| |
| − | Postgres Database name: V.cholerae_20151027_gmb3build5
| |
| − |
| |
| − | UniProt XML filename: [[Media:uniprot-organism_BL_20151027.xml.gz|compressed uniprot-organism%3A243277_BL_20151027.xml]]
| |
| − | * UniProt XML version (found out using the [http://www.uniprot.org/help/?fil=section:news UniProt News Page]: UniProt release 2015_10
| |
| − | * UniProt XML download link: http://www.uniprot.org/uniprot/?query=organism:243277#
| |
| − | * Time taken to import: 2.99 minutes
| |
| − | ** Note: Import times were as expected; nothing unusual occurred during the import process.
| |
| − |
| |
| − | GO OBO-XML filename: [[Media:BL 20151027 go daily-termdb.zip|compressed BL_20151027_go_daily-termdb.obo-xml]]
| |
| − | * GO OBO-XML version (derived from the date modified on the file, itself): Date modified - ''10/27/2015 2:24 AM''
| |
| − | * GO OBO-XML download link: [http://geneontology.org/page/download-ontology#Legacy_Downloads GO website]
| |
| − | * Time taken to import: 6.98 minutes
| |
| − | * Time taken to process: 4.54 minutes
| |
| − | ** Note: Import times were as expected; nothing unusual occurred during the import process.
| |
| − |
| |
| − | GOA filename: [[Media:46.V_cholerae_ATCC_39315_BL_20151027.zip| compressed 46.V_cholerae_ATCC_39315_BL_20151027.goa]]
| |
| − | * GOA version (taken from [http://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/ here]: Last Update - ''13-Oct-2015 07:31''
| |
| − | * GOA download link: [http://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/46.V_cholerae_ATCC_39315.goa link]
| |
| − | * Time taken to import: 0.06 minutes
| |
| − | ** Note: Import times were as expected; nothing unusual occurred during the import process.
| |
| − |
| |
| − | Name of .gdb file: [[Media:Vc-Std_20151027_BL.zip | compressed Vc-Std_20151027_BL.gdb]]
| |
| − | * Time taken to export: one hour and nineteen minutes
| |
| − | ** Start time: 3:56
| |
| − | ** End time: 5:15
| |
| − | Note: Export times were as expected; nothing unusual occurred during the export process. Program windows, however, were closed by the time the export product was observed; the time taken to export was found out by observing the time on the Date Modified section of the produced file. The found end time was in line with the times found by others (ranged around an hour and 15 minutes)
| |
| − |
| |
| − | ===Using TallyEngine to test the newly created database===
| |
| − | *PostgreSQL was initialized through pgAdmin III and the database V.cholerae_20151027_gmb3build5 was left running
| |
| − | *GenMAPP builder was booted and ''Run XML and Database Tallies for UniProt and GO'' was selected under the ''Tallies'' menu item; the UniProt XML and GO files that were imported were chosen
| |
| − | *'''Results of TallyEngine:'''
| |
| − | *[[File:TallyEngine_Results_BL_20151029.png]]
| |
| − |
| |
| − | ===Using XMLPipeDB match to Validate the XML Results from TallyEngine===
| |
| − | *The Windows command line was launched (cmd.exe)
| |
| − | *This set of commands was inputted into the command line in order to utilize XMLPipeDB match:
| |
| − | <code>java -jar xmlpipedb-match-1.1.1.jar "VC_[0-9][0-9][0-9][0-9]" < uniprot-organism%3A243277.xml</code>
| |
| − | **NOTE: Prior to executing the command, the folder that held the files and xmlpipedb-match-1.1.1.jar was entered through the Windows command line (a set of CD commands was used in order to enter the correct directory)
| |
| − | *2738 unique matches were found through XMLPipeDB match
| |
| − | **[[File:BL MATCHUTILITY WEEK9.png]]
| |
| − | Are your results the same as you got for the TallyEngine? Why or why not?
| |
| − | *These results are not the same as what was found using TallyEngine (3831 results were found through TallyEngine). It is suspected that the match command pattern is not fully representative of everything that falls under ''OrderedLocusNames'' (the fault, it is believed, lies with match and not with TallyEngine)
| |
| − |
| |
| − | ===Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine===
| |
| − | *''pgAdmin III'' was booted and all of the necessary connections were made
| |
| − | *It was realized that the ''gene/name'' tags in the XML file end up in the ''genenametype'' table (source: [[How_Do_I_Count_Thee%3F_Let_Me_Count_The_Ways |the wiki page regarding database quality analysis]]
| |
| − | *In ''pgAdmin III'', the query <code>select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_[0-9][0-9][0-9][0-9]';</code> was issued via the SQL Query menu in order to further validate the PostgreSQL Database Results according to TallyEngine
| |
| − | *2737 unique matches were found in ''pgAdmin III'' (postgres database results). These results are not the same as what was found by TallyEngine and are slightly different from what was found using XMLPipeDB match (1 off). It seems that the results are really close to what the Match utility found because it utilizes the same pattern ('VC_[0-9][0-9][0-9][0-9]')
| |
| − | **[[File:BL PSQL COUNT WEEK9.png]]
| |
| − |
| |
| − | ===OriginalRowCounts Comparison using Microsoft Access===
| |
| − | *The exported ''Vc-Std_20151027_BL.gdb'' file was opened using Microsoft Access
| |
| − | *7664 unique matches found using Microsoft Access were found by observing the row count for the ''OrderedLocusNames'' table. This number of unique matches is very different from what was found using the other methods (more than double of what was found using TallyEngine, which is 3831 unique results). It is believed that the ''OrderedLocusNames'' table contains two sets of data (different "versions") with each harboring one value/ID that is unaccounted for by the other count validation methods.
| |
| − |
| |
| − | ===Visual Inspection===
| |
| − | ====Observing the 'OrderedLocusNames' table, and further modifications====
| |
| − | *The database was opened in a program that is able to read .mdb files, such as Microsoft Access
| |
| − | *Looking at the OrderedLocusNames column, it was noticed that some of the entries contain underscores after the ''VC'' and others do not. By filtering so that only the entries with underscores appear, it was noticed that there are 3832 total entries with underscores. 3832 is half of 7664, which is the total amount of entries in the OrderedLocusNames table; this suggests that there exists two different versions of the data (with underscores and without). It was also noticed that both versions of the data (underscores and without) have some entries that include a letter A after the ''VC'' portion; through filtering, it was noticed that the entries with A's account for 1095 of the entries in both sets of data.
| |
| − | *Including the As in the command for the Match utility, <code>java -jar xmlpipedb-match-1.1.1.jar "VC_A?[0-9][0-9][0-9][0-9]" < uniprot-organism%3A243277.xml</code>, resulting in a result of 3831 unique matches; 3831 is an exact match of what was found using TallyEngine.
| |
| − | *Modifying the SQL query so that the pattern includes the entries with A's, by using <code>select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_A?[0-9][0-9][0-9][0-9]';</code>, showed that there are 3831 entries (the number is in line with what TallyEngine found).
| |
| − | *It now appears that there is one entry in both data sets (underscore and non-underscore) that remains unaccounted for
| |
| − | ===Using Microsoft Excel to Compare ID Lists===
| |
| − | *The 'OrderedLocusNames' table was first put in ascending order (lower ID #s come first) and was then exported in a format that Excel can read (was then opened with Excel)
| |
| − | *<code>java -jar xmlpipedb-match-1.1.1.jar "VC_A?[0-9][0-9][0-9][0-9]" < uniprot-organism%3A243277.xml > ordered_locus_match_results_BL.txt</code> was used via the Windows command line in order to export the ID lists with the underscores to a text file (one half of all of the IDs)
| |
| − | *The Match utility exported text file was opened via Excel and the colon ('':'') was set as the delimiter (since a colon separates the match count from the ID)
| |
| − | *pgAdmin III was booted up and the proper database was selected (V.cholerae_20151027_gmb3build5); the SQL query <code>select value from genenametype where type = 'ordered locus';</code> was inputted and "Execute query, write result to file" was clicked. Through these steps, the list of IDs from PostgreSQL was exported in an Excel friendly format (such as .csv)
| |
| − | *The column of IDs from 'OrderedLocusNames', the one exported from the Match utility, and the one that was exported from PostgreSQL, were put side by side in a new Excel document with no spaces between them; it was ensured that each column was in ascending order. The column of IDs from 'OrderedLocusNames' was given the label ' 'IDs FROM 'ORDEREDLOCUSNAMES' ', the one that was exported from the Match utility was given the label ''IDs FROM MATCH'', and the one that was exported from PostgreSQL was given the label ''IDs FROM postgreSQL''
| |
| − | *A third column was inserted directly to the right of the last column with the heading ''MATCH''
| |
| − | *''IDs FROM 'ORDEREDLOCUSNAMES'' was additionally labeled with a number 1, ''IDs FROM MATCH'' was additionally labeled with a number 2, and ''IDs FROM postgreSQL'' was additionally labeled with a number 3
| |
| − | *6 empty adjacent columns were reserved for Excel MATCH commands (which look up ID values from one column in another column/range); MATCH outputs the position where the matched value was found (starting from 1) or '''#N/A''' if there existed no match at that position
| |
| − | *6 empty columns were labeled directly to the right (adjacent) to ''IDs FROM postgreSQL (3)''; in order, from left to right, the column labels typed in were ''MATCH:1 to 2'', ''MATCH:1 to 3'', ''MATCH:2 to 1'', ''MATCH:2 to 3'', ''MATCH:3 to 1'', and ''MATCH:3 to 2''
| |
| − | **NOTE: In the MATCH column labels, the numbers 1, 2, and 3 correspond to distinct ID columns; 1 represents ''IDs FROM 'ORDEREDLOCUSNAMES'', 2 represents ''IDs FROM MATCH'', and 3 represents ''IDs FROM postgreSQL''
| |
| − | *MATCH commands were then written for each of the 6 MATCH columns and applied to the entirety of each MATCH column; the basic format is <code>=MATCH(VALUE TO LOOK-UP, RANGE/COLUMN WHERE THE LOOKING-UP OF A VALUE TAKES PLACE, "MATCH TYPE" [0 in this case])</code>. The purpose of these MATCH commands is to compare the three different ID lists (with each other)
| |
| − |
| |
| − | '''ALL MATCH COMMANDS:'''
| |
| − | Format - '''Column Label''' : <code>MATCH Command</code> (in first "cell")
| |
| − | '''MATCH:1 to 2''' : <code>=MATCH(A2, B$2:B$3832,0)</code>
| |
| − | '''MATCH:1 to 3''' : <code>=MATCH(A2, C$2:C$3832,0)</code>
| |
| − | '''MATCH:2 to 1''' : <code>=MATCH(B2, A$2:A$3833,0)</code>
| |
| − | '''MATCH:2 to 3''' : <code>=MATCH(B2, C$2:C$3832,0)</code>
| |
| − | '''MATCH:3 to 1''' : <code>=MATCH(C2, A$2:A$3833,0)</code>
| |
| − | '''MATCH:3 to 2''' : <code>=MATCH(C2, B$2:B$3832,0)</code>
| |
| − | ====MATCH results analysis====
| |
| − | *''ctrl+F'' was utilized in order to find instances of '''#N/A''' among all of the MATCH columns (some options included, Look In: ''Values'', Find What: ''#N/A'', Find All); eight cases of '''#N/A''' were found
| |
| − | *By observing the cases of '''#N/A''' (and the corresponding IDs), it was realized that 6 of the cases were due to the presence of '''VC_1738/VC_1739''' in position '''1711''' within the ''IDs FROM postgreSQL (3)'' column
| |
| − | *The last two cases of '''#N/A''' were found to be the result of the presence of '''VC_A0360.1''' in position '''3093''' within the ''IDs FROM 'ORDEREDLOCUSNAMES (1)'' column, and in position '''3092''' within the ''IDs FROM postgreSQL (3)'' column
| |
| − | *It seems apparent that there exist some entries in the 'ORDEREDLOCUSNAMES' that are not accounted for by the MATCH utility ('''VC_A0360.1'''); it is also clear that there are two entries acting as one entry within the postgreSQL output ('''VC_1738/VC_1739'''). '''VC_A0360.1''' also exists within the postgreSQL output.
| |
| − | ===Further Quality Assurance Work===
| |
| − | *Using the Match utility, <code>java -jar xmlpipedb-match-1.1.1.jar "VC_A?[0-9][0-9][0-9][0-9](.1)?" < uniprot-organism%3A243277.xml</code> was inputted into the Windows command line in order to get all 3832 possible unique matches (includes '''VC_A0360.1''')
| |
| − | *In pgAdmin III, the SQL query <code>select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_A?[0-9][0-9][0-9][0-9](.1)?';</code> returns the same count as the previously attempted query (the previously attempted query appears to have taken in account to the presence of '''VC_A0360.1'''); the issue that is preventing a count of 3832 seems to be the presence of '''VC_1738/VC_1739''' in one of the rows:
| |
| − | **[[File:Pgsql_issuerow_BL_wk9.png]]
| |
| − | ==.gdb Use in GenMAPP==
| |
| − | *Some of the protocol from [http://www.openwetware.org/wiki/BIOL367/F10:GenMAPP_and_MAPPFinder_Protocols Part 2 of the ''Vibrio cholerae'' Microarray Data Analysis] was used as a reference for this portion of the assignment
| |
| − | *''Vc-Std_20151027_BL.gdb'', the recently created ''Vibrio cholerae'' database, was placed within the Gene Databases folder of the GenMAPP directory (the folder is within the GenMAPP 2 Data folder)
| |
| − | *GenMAPP (Version 2.1) was launched
| |
| − | *The new gene database was loaded by going into ''Data > Choose Gene Database''
| |
| − | *The tab deliminated GenMAPP formatted [[Media:Merrell_Compiled_Raw_Data_Vibrio_BL_20151015.txt|data]] from week 8 was loaded into GenMAPP through ''Data > Expression Dataset Manager > Expression Datasets > New Dataset > Merrell_Compiled_Raw_Data_Vibrio_BL_20151015.txt''
| |
| − |
| |
| − |
| |
| − |
| |
| − | While the above sections perform quality assurance on the exported Gene Database via verifying ID counts, the "proof in the pudding" is to actually use the Gene Database in GenMAPP. You can follow the instructions in [http://www.openwetware.org/wiki/BIOL367/F10:GenMAPP_and_MAPPFinder_Protocols Part 2 of the ''Vibrio cholerae'' Microarray Data Analysis] to verify that the Gene Database works in GenMAPP. In this case, the emphasis is not on the findings of the data analysis itself, but that the Gene Database functions appropriate in GenMAPP.
| |
| − |
| |
| − | Note:
| |
| − |
| |
| − | ===Putting a gene on the MAPP using the GeneFinder window===
| |
| − |
| |
| − | * Try a sample ID from each of the gene ID systems. Open the Backpage and see if all of the cross-referenced IDs that are supposed to be there are there.
| |
| − |
| |
| − | Note:
| |
| − |
| |
| − | ===Creating an Expression Dataset in the Expression Dataset Manager===
| |
| − |
| |
| − | * How many of the IDs were imported out of the total IDs in the microarray dataset? How many exceptions were there? Look in the EX.txt file and look at the error codes for the records that were not imported into the Expression Dataset. Do these represent IDs that were present in the UniProt XML, but were somehow not imported? or were they not present in the UniProt XML?
| |
| − |
| |
| − | Note:
| |
| − |
| |
| − | ===Coloring a MAPP with expression data===
| |
| − |
| |
| − | Note:
| |
| − |
| |
| − | ===Running MAPPFinder===
| |
| − |
| |
| − | Note:
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − |
| |
| − | ===OriginalRowCounts Comparison===
| |
| − |
| |
| − | Within the .gdb file, look at the OriginalRowCounts table to see if the database has the expected tables with the expected number of records. Compare the tables and records with a benchmark .gdb file.
| |
| − |
| |
| − | Benchmark .gdb file:
| |
| − |
| |
| − |
| |
| − | ----
| |
| − | {{Template:blitvak}}
| |
| | ==Initial Preparations== | | ==Initial Preparations== |
| | ''List of required programs was initially sourced from the [[Running GenMAPP Builder | Running GenMAPP Builder page]]'' | | ''List of required programs was initially sourced from the [[Running GenMAPP Builder | Running GenMAPP Builder page]]'' |
In preparation for this assignment, it was ensured that these programs were installed on a Windows workstation:
Note: Export times were as expected; nothing unusual occurred during the export process. Program windows, however, were closed by the time the export product was observed; the time taken to export was found out by observing the time on the Date Modified section of the produced file. The found end time was in line with the times found by others (ranged around an hour and 15 minutes)