Difference between revisions of "Kevin Wyllie Week 15"
(Added the link for our final presentation.) |
(Added the statement of work.) |
||
| Line 41: | Line 41: | ||
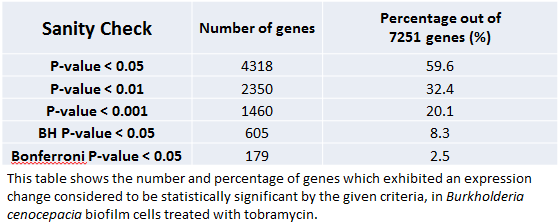
[[file:KWVPSanitycheck2.png|center]] | [[file:KWVPSanitycheck2.png|center]] | ||
| + | |||
| + | |||
| + | == Reflection == | ||
| + | |||
| + | === Statement of Work === | ||
| + | |||
| + | The goal of this project was to create a gene database for ''Burkholderia cenocepacia'' and to use this database for the purpose of conducting an analysis on microarray data generated by Van Acker et al (2013). This data measured the transcriptomic effects of tobramycin (an antibiotic) on the bacterium. The database was constructed using data from [http://www.burkholderia.com/ Burkholderia Genome Database], [http://www.uniprot.org/ UniProt] and [http://geneontology.org/ Gene Ontology Consortium]. The microarray data was taken from the paper, [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3596321/ Biofilm-Grown ''Burkholderia cepacia'' Complex Cells Survive Antibiotic Treatment by Avoiding Production of Reactive Oxygen Species]. Once the gene database (found [[Media:Bc-Std_GEN_20151204.zip|here]]) was created, the processed microarray data (found [[media:Raw_compiled_data_FINAL_KWVP20151507.xlsx|here]]) were fed through a software called GenMAPP, which analyzed the data and returned significantly changed cellular processes (gene ontology terms - see [[Kevin Wyllie Week 15|week 15 journal]] for files and results). We then compared the biological implications of our analysis to that of Van Acker et al (2013). | ||
| + | |||
| + | === Assessment of Project === | ||
| + | |||
| + | * Give an objective assessment of the success of your project workflow and teamwork. | ||
| + | * What worked and what didn't work? | ||
| + | * What would you do differently if you could do it all over again? | ||
| + | * Evaluate the Gene Database Project and Group Report in the following areas: | ||
| + | *# Content: What is the quality of the work? | ||
| + | *# Organization: Comment on the organization of the project and of your group's wiki pages. | ||
| + | *# Completeness: Did your team achieve all of the project objectives? Why or why not? | ||
| + | |||
| + | === Reflection on the Process === | ||
| + | |||
| + | * What did you learn? | ||
| + | ** With your head (biological or computer science principles) | ||
| + | ** With your heart (personal qualities and teamwork qualities that make things work or not work)? | ||
| + | ** With your hands (technical skills)? | ||
| + | * What lesson will you take away from this project that you will still use a year from now? | ||
== Links == | == Links == | ||
Revision as of 05:30, 15 December 2015
Contents
Final Presentation
GENialOMICS Final Presentation
Electronic Lab Notebook
MAPPFinder Procedure
- MAPPFinder was launched from the GenMAPP window: "Tools" > "MAPPFinder".
- "Calculate New Results" was selected from the MAPPFinder window.
- "Find File" was selected and in the directory, the .gex file (created in the Expression Dataset Manager - see week 14) was selected.
- This file can be found here.
- Under the "Select Color Set" field, "KWVP_20151205" was selected (the color set's name in this case is the same as the .gex file's name - and there's only one color set in the .gex file).
- After the color set has been selected, under the "Select Criteria to filter by" field, either increase or decrease was chosen.
- The boxes for "Gene Ontology" and "Click here to calculate p values..." were checked.
- The location to save the resulting .txt file was selected with the "Browse" option toward the bottom of the window. Note that because this protocol was carried out both for increase and decrease, there were two text files generated in MAPPFinder.
- "Run MAPPFinder" was selected to generate the gene ontology tree (and the aforementioned .txt files).
- The files generated can be found here:
Filtered GO List
- The GO term files above were opened in Excel, and the following filters were placed on the given columns:
- Z score: greater than 2
- PermuteP: less than 0.05
- Number changed: greater than or equal to 4 AND less than 100
- Percent changed: great than or equal to 15
- Values in the following columns were recorded:
- Number Changed
- Number Measured
- Number in GO
- Percent Changed
- Percent Present
- PermuteP
- AdjustedP
- Some of the above filter criteria had to be adjusted to attain 16 non-redundant GO terms. A list of these terms is shown below. For those terms which did not exactly fit this initial criteria, the altered criteria is apparent in the values on the table.
Sanity Check
Reflection
Statement of Work
The goal of this project was to create a gene database for Burkholderia cenocepacia and to use this database for the purpose of conducting an analysis on microarray data generated by Van Acker et al (2013). This data measured the transcriptomic effects of tobramycin (an antibiotic) on the bacterium. The database was constructed using data from Burkholderia Genome Database, UniProt and Gene Ontology Consortium. The microarray data was taken from the paper, Biofilm-Grown Burkholderia cepacia Complex Cells Survive Antibiotic Treatment by Avoiding Production of Reactive Oxygen Species. Once the gene database (found here) was created, the processed microarray data (found here) were fed through a software called GenMAPP, which analyzed the data and returned significantly changed cellular processes (gene ontology terms - see week 15 journal for files and results). We then compared the biological implications of our analysis to that of Van Acker et al (2013).
Assessment of Project
- Give an objective assessment of the success of your project workflow and teamwork.
- What worked and what didn't work?
- What would you do differently if you could do it all over again?
- Evaluate the Gene Database Project and Group Report in the following areas:
- Content: What is the quality of the work?
- Organization: Comment on the organization of the project and of your group's wiki pages.
- Completeness: Did your team achieve all of the project objectives? Why or why not?
Reflection on the Process
- What did you learn?
- With your head (biological or computer science principles)
- With your heart (personal qualities and teamwork qualities that make things work or not work)?
- With your hands (technical skills)?
- What lesson will you take away from this project that you will still use a year from now?
Links
| Weekly Group Assignments | Shared Group Journals | Project Links | Team Members |
|---|---|---|---|
|
|
|
|
|
- Kevin Wyllie Week 2 (See the original assignment and class journal.)
- Kevin Wyllie Week 3 (See the original assignment and class journal.)
- Kevin Wyllie Week 4 (See the original assignment and class journal.)
- Kevin Wyllie Week 5 (See the original assignment and class journal.)
- Kevin Wyllie Week 6 (See the original assignment and class journal.)
- Kevin Wyllie Week 7 (See the original assignment and class journal.)
- Kevin Wyllie Week 8 (See the original assignment and class journal.)
- Kevin Wyllie Week 9 (See the original assignment and class journal.)
- Kevin Wyllie Week 10 (See the original assignment.)
- Kevin Wyllie Week 11 (See the original assignment.)
- Kevin Wyllie Week 12 (See the original assignment.)
- Kevin Wyllie Week 14 (See the original assignment.)
- Kevin Wyllie Week 15 (See the original assignment.)