Kevin Wyllie Week 4
Contents
Transcription and Translation “Taken to the Next Level”
Question 1
cat infA-E.coli-K12.txt | sed -r "s/tt[gt]ac[at](.){17}[ct]at[at]at/ <minus35box>&<\/minus10box> /g"
The sed command shown finds the beginning tag for the -35 box and the end tag for the -10 box, based on their consensus sequence and known distance from each other...
cat infA-E.coli-K12.txt | sed -r "s/tt[gt]ac[at](.){17}[ct]at[at]at/ <minus35box>&<\/minus10box> /g" | sed "s/<minus35box>tt[gt]ac[at]/&<\/minus35box> /g" | sed "s/[ct]at[at]at<\/minus10box>/ <minus10box>&/g"
...and these sed commands add the remaining tags for the -35 and -10 box, based on the location of either existing tag.
cat infA-E.coli-K12.txt | ... | sed -r "s/[ct]at[at]at<\/minus10box>(.){6}/& <tss>/g" | sed "s/<tss>./&<\/tss> /g"
These commands add the tags for the transcription start site (TSS), based on its consensus sequence and known distance from the -10 box.
cat infA-E.coli-K12.txt | ... | sed "s/gagg/ <rbs>&<\/rbs> /g"
This sed command adds the ribosome binding site (RBS), based its consensus sequence.
cat infA-E.coli-K12.txt | ... | sed "s/aaaaggt/ <terminator>&/g" | sed "s/gcctttt…./&<\/terminator> /g"
This sed command adds the tags for the terminator, based off of its consensus sequence and hairpin behavior.
cat infA-E.coli-K12.txt | ... | sed "s/<\/rbs> /&\n/g" | sed "2s/atg/ <start_codon>&<\/start_codon> /1"
These commands start a new line after the RBS and then add start-codon tags to the first occurring "atg" sequence on the newly separated line.
cat infA-E.coli-K12.txt | ... | sed "2s/ta[ag]/\n&/g" | sed "3,10s/tga/\n&/g"
These commands start a new line for each potential stop codon sequence occurring after the RBS.
cat infA-E.coli-K12.txt | ... | sed "17s/tga/ <stop_codon>&<\/stop_codon> /g"
These commands at stop-codon tags to the potential stop codon sequence occurring closest to (but not after) the terminator.
cat infA-E.coli-K12.txt | ... | sed ':a;N;$!ba;s/\n//g'
This rather unintelligible command undoes any line breaks, combining the text file back into one line.
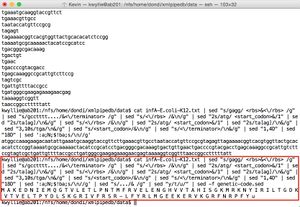
- The final pipe is shown entered into the command line, along with the output. The tagged sequence is:
ttttcaccacaagaatgaatgttttcggcacatttctccccagagtgttataattgcggtcgcagagttggttacgctcattaccccgctgccgataa ggaatttttcgcgtcaggtaacgcccatcgtttatctcaccgctcccttatacgttgcgcttttggtgcggcttagccgtgtgttttcggagtaatgtgc cgaacctgtttgttgcgatttagcgcgcaaatc <minus35box>tttact</minus35box> tatttacagaacttcgg <minus10box> cattat</minus10box> cttgc <tss>c</tss> ggttcaaattacggtagtgatacccca <rbs>gagg</rbs> attag <start_codon>atg</start_codon> gccaaagaagacaatattgaaatgcaaggtaccgttcttgaaacgttgcctaataccatgttc cgcgtagagttagaaaacggtcacgtggttactgcacacatctccggtaaaatgcgcaaaaactacatccgcatcctgacgggcgacaaagtg actgttgaactgaccccgtacgacctgagcaaaggccgcattgtcttccgtagtcgctgattgttttaccgcc <stop_codon>tga</stop_codon> tgggcgaagagaaagaacgagt <terminator>aaaaggtcggtttaaccggcctttttatt </terminator> ttat
Question 2
The mRNA sequence can be isolated from the file using some of the same commands as before.
sed -r "s/tt[gt]ac[at](.){17}[ct]at[at]at/ <minus35box>&<\/minus10box> /g" | sed "s/<minus35box>tt[gt]ac[at]/&<\/minus35box> /g" | sed "s/[ct]at[at]at<\/minus10box>/ <minus10box>&/g" | sed -r "s/[ct]at[at]at<\/minus10box>(.){6}/& <tss>/g"
The pipe starts wiith question 1's first few commands, up to the point of adding the TSS tags. However, the TSS end tag has been omitted.
sed -r "s/tt[gt]ac[at](.){17}[ct]at[at]at/ <minus35box>&<\/minus10box> /g" | ... | sed "s/gcctttt..../&<\/terminator> /g"
This command adds the terminator's end tag, but not its beginning tag.
sed -r "s/tt[gt]ac[at](.){17}[ct]at[at]at/ <minus35box>&<\/minus10box> /g" | ... | sed "s/<tss>/&\n/g" | sed "s/<\/terminator>/\n&/g"
These commands begin new lines after the TSS beginning tag and before the terminator's end tag.
sed -r "s/tt[gt]ac[at](.){17}[ct]at[at]at/ <minus35box>&<\/minus10box> /g" | ... | sed "3D" | sed "1D"
These commands delete the top and bottom (first and third) lines, leaving only the middle.
- The final pipe is shown entered into the command line, along with the output. The transcribed sequence is:
cggttcaaattacggtagtgataccccagaggattagatggccaaagaagacaatattgaaatgcaaggtaccgttcttgaaacgttgcctaataccatgttccgcgtagagttagaaaacggtcacg tggttactgcacacatctccggtaaaatgcgcaaaaactacatccgcatcctgacgggcgacaaagtgactgttgaactgaccccgtacgacctgagcaaaggccgcattgtcttccgtagtcgctg attgttttaccgcctgatgggcgaagagaaagaacgagtaaaaggtcggtttaaccggcctttttatt
Question 3
Again, the amino acid sequence can be found by using modified bits and pieces from question 1.
- The pipe is shown entered into the command line, along with the output. The amino sequence this returns is:
M A K E D N I E M Q G T V L E T L P N T M F R V E L E N G H V V T A H I S G K M R K N Y I R I L T G D K V T V E L T P Y D L S K G R I V F R S R - L F Y R L M G E E K E R V K G R F N R P F Y
Protcol
Protocol - Question 1
- The trickiest part of this assignment is the first part. You must label both the -35 and -10 sequences with beginning and end tags, however, the consensus sequences given may appear multiple times in the text file, and only one of them is the correct location. This can be solved via the given information that the -35 and -10 boxes are 17 bases apart. Thus, between the -35 end tag and the -10 beginning tag, there should be 17 bases.
-
sed -r "s/tt[gt]ac[at](.){17}[ct]at[at]at/ <minus35box>&<\/minus10box> /g" - The
-ris added to the sed command because of the syntax used in the query. The(.){17}tells the computer that there should be 17 characters between the queries on either side of this syntax.
-
- Now that you can be sure that these two tags are in the right place, use their locations to add the remaining two ( the -35 beginning tag and the -10 end tag).
-
sed "s/<minus35box>tt[gt]ac[at]/&<\/minus35box> /g" | sed "s/[ct]at[at]at<\/minus10box>/ <minus10box>&/g"
-
- Since the transcription start sight is the 12th base from the beginning of the -10 box, and the -10 box itself is 6 bases long, the TSS should be 6 bases from the -10 end tag.
-
sed -r "s/[ct]at[at]at<\/minus10box>(.){6}/& <tss>/g" | sed "s/<tss>./&<\/tss> /g"
-
- Now find and tag the ribosome binding site (RBS).
-
sed "s/gagg/ <rbs>&<\/rbs> /g"
-
- Next is the terminator. You're given the initial hairpin sequence, and can use the base pairing rules (along with the information given about how this sequence breaks these rules slightly) to determine the end sequence.
-
sed "s/aaaaggt/ <terminator>&/g" | sed "s/gcctttt…./&<\/terminator> /g" - The second hairpin sequence is followed by four "wildcards" because you're given that there are four characters following this sequence which are still part of the terminator sequence.
-
- The next step is to tag the start codon. However, there are likely multiple
atgsequences in this gene. So, you should assume that the true start codon is closest to (but still after) the RBS. To do this, you must separate lines.-
sed "s/<\/rbs> /&\n/g" - This places a new line after the RBS end tag.
-
- Now that a new line has been formed, tag the first
ATGsequence on this line.-
sed "2s/atg/ <start_codon>&<\/start_codon> /1" - The
2stells sed to only look at the second line, while the1tells sed to only replace the first match in the line.
-
- Labelling the stop codons is equally as tough as doing so for the start codons, and for the same reason. So you must create more line breaks, at each potential sequence. And assume that the true stop codon is the one closest to (but still before) the terminator.
-
sed "2s/ta[ag]/\n&/g" | sed "3,10s/tga/\n&/g" -
sed "17s/tga/ <stop_codon>&<\/stop_codon> /g"
-
- Now just undo the line breaks.
-
sed ':a;N;$!ba;s/\n//g'
-
Protocol - Question 2
Bits and pieces of the previous pipe can be used to find the mRNA strand.
- Start by beginning with the previous pipe, up until addition of the TSS tags. At this point, add the beginning tag, but not the end tag.
-
sed -r "s/[ct]at[at]at<\/minus10box>(.){6}/& <tss>/g"
-
- Now add the end tag (but not the beginning tag) of the terminator sequence.
-
sed "s/gcctttt..../&<\/terminator> /g"
-
- Then start new lines after the TSS beginning tag and before the terminator end tag.
- sed "s/<tss>/&\n/g" | sed "s/<\/terminator>/\n&/g"
- Now delete lines 1 and 3 to isolate the mRNA sequence.
- sed "3D" | sed "1D"
- The
3Dand1Ddesignate that lines 3 and 1 should be deleted.
Protocol - Question 3
- As done for question 2, use the appropriate modified pieces of the pipe from question 1 to find the amino acid sequence.
- Once the translated sequence has been isolated, use the translation pipe from the week 3 assignment.
Links
- Kevin Wyllie Week 2 (See the original assignment and class journal.)
- Kevin Wyllie Week 3 (See the original assignment and class journal.)
- Kevin Wyllie Week 4 (See the original assignment and class journal.)
- Kevin Wyllie Week 5 (See the original assignment and class journal.)
- Kevin Wyllie Week 6 (See the original assignment and class journal.)
- Kevin Wyllie Week 7 (See the original assignment and class journal.)
- Kevin Wyllie Week 8 (See the original assignment and class journal.)
- Kevin Wyllie Week 9 (See the original assignment and class journal.)
- Kevin Wyllie Week 10 (See the original assignment.)
- Kevin Wyllie Week 11 (See the original assignment.)
- Kevin Wyllie Week 12 (See the original assignment.)
- Kevin Wyllie Week 14 (See the original assignment.)
- Kevin Wyllie Week 15 (See the original assignment.)