Kevin Wyllie Week 9
Contents
- 1 Pre-requisites
- 2 Download and Extract Data Source Files
- 3 Export Information
- 4 Download or Update GenMAPP Builder
- 5 Create New Database in PostgreSQL
- 6 Configuring GenMAPP Builder to Connect to your PostgreSQL Database
- 7 Importing Data into the PostgreSQL Database
- 8 Exporting a GenMAPP Gene Database (.gdb)
- 9 Checking the Quality of your Exported Gene Database
- 10 TallyEngine
- 11 Using XMLPipeDB match to Validate the XML Results from the TallyEngine
- 12 Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine
- 13 OriginalRowCounts Comparison
- 14 Visual Inspection
- 15 .gdb Use in GenMAPP
- 16 Compare Gene Database to Outside Resource
- 17 Links
Pre-requisites
This protocol assumes that you are working in a Windows environment. While it is possible to run GenMAPP Builder under the Mac or Linux OS, the end product, a GenMAPP-compatible Gene Database (.gdb), can only be used with the GenMAPP program, which can only be run on Windows. This set of software has already been installed on the computers in the Seaver 120 computer lab. If you want to perform this procedure on your own machine, you must set up your working environment with:
- Any tool that can unpack .gz and .zip files
- We use 7-zip
- Note that we have found that the native Windows utility cannot reliably unpack .gz files or .zip files containing .jar files.
- PostgreSQL on Windows (http://www.enterprisedb.com/products-services-training/pgdownload)
- This tutorial was written using PostgreSQL 9.4.x.
- GenMAPP Builder (https://sourceforge.net/projects/xmlpipedb/files/)
- Java JDK 1.8 64-bit
- Download page
- File to download is: jdk-8u65-windows-x64.exe
- GenMAPP 2 can be downloaded here. The file to download is "GenMAPPv2Setup.exe".
- XMLPipeDB match utility (https://sourceforge.net/projects/xmlpipedb/files/) for counting IDs in XML files
- Microsoft Access or any other tool that can read .mdb files
Download and Extract Data Source Files
- Download UniProt XML, GOA, and GO OBO-XML files.
UniProt XML
- The UniProt XML file was found on the UniProt Complete Proteomes page.
- "Bacteria" was selected, under the "Superkingdom" heading (on left), followed by "Reference proteome."
- After scrolling through results, Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961) was found.
- Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961) was selected, followed by the "download" button at the top of the next page. This opened a dialogue box:
- "Download all" was selected.
- "XML" was selected from the "Format" drop down menu.
- "Compressed" format was chosen.
GOA
- This is the UniProt-GOA home page.
- The current UniProt-GOA file was downloaded from the UniProt-GOA ftp site.
- In the directory that appeared, "proteomes" directory was chosen.
- This paged took several minutes to load.
- Vibro cholerae was right-clicked, and "save link as" was selected.
- Left-clicking on this link prompted the file to open in a browser window.
- The version information was found in the ftp file directory under the "Last modified" column.
GO OBO-XML
- Download the GO OBO-XML formatted file from the Gene Ontology download page. Click on the link for "obo-xml.gz" under the heading "Legacy Downloads."
- This file is updated daily. You can get the day/time that the file was created from the file properties after you have unzipped the file.
Extract the UniProt XML and GO OBO-XML files
- The UniProt XML and GO OBO-XML files were extracted using 7-zip.
Export Information
Version of GenMAPP Builder: 5
Computer on which export was run: HP Compaq 8300 Elite SFF FC
Postgres Database name: pgAdminIII
UniProt XML filename: uniprot-organism%3A243277
- UniProt XML version: 2015_10
- UniProt XML download link: http://www.uniprot.org/uniprot/?query=organism:243277
- Time taken to import: 3.02 minutes
GO OBO-XML filename: go_daily-termdb.obo-xml
- GO OBO-XML version (The version information can be found in the file properties after the file downloaded from the: 2015
- GO OBO-XML download link: http://geneontology.org/page/download-ontology#Legacy_Downloads
- Time taken to import: 7.68 minutes
- Time taken to process: 4.48
GOA filename: 46.V_cholerae_ATCC_39315.goa
- GOA version: 2015
- GOA download link: http://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes/46.V_cholerae_ATCC_39315.goa
- Time taken to import: 0.06
Name of .gdb file: Vc-Std_20151029_KW2.gdb
- Time taken to export:
- Start time: 2:40pm
- End time: 3:52pm
Download or Update GenMAPP Builder
- gmbuilder-3.0.0-build-5.zip was downloaded from the XMLPipeDB releases page on GitHub.
- The GenMAPP Builder folder was extracted using 7-zip.
- Move the folder to a safe storage domain.
Create New Database in PostgreSQL
NOTE: if you have already performed this step and want to use GenMAPP Builder functions with a database you previously created in PostgreSQL, you can skip this step.
- Launch pgAdmin III.
- Double-click on PostgreSQL 9.4 (localhost:5432) on the upper left hand side of the window.
- This is the equivalent of connecting you to the server and you may be asked for a password at this point.
- Right click on "Databases" and Select "New Database..."
- Give the database a name in the "Name" field and click OK.
- Take some care in selecting a meaningful name. It is good practice to at least include the species and today's date in the name.
- Double-left-click on your new database name in the treeview on the left.
- Click on the SQL icon in the toolbar at the top of the window.
- The SQL Editor tab will be open and there may be leftover query text in the upper pane. Delete this text. You are now going to use an XMLPipeDB query to create the tables in the database.
- Click on the Open File icon in the toolbar (the yellow folder with an arrow).
- Navigate to the folder in which you unzipped GenMAPP Builder.
- Open the sql folder and open the file gmbuilder.sql. You should see SQL code appear in the SQL Editor tab.
- Click the Execute Query icon which looks like a green "Play" triangle button.
- You should get a series of NOTICE messages in the Messages tab at the bottom of the window, concluding with a message like "Query returned successfully with no result in 15583 ms" in the end. This query now created all the tables in the database (although there is still no data in them).
- Close the query window (you don't need to save the query because you have already run it).
- To double check that all is OK, click the + sign for the database, then the + sign for Schemas, then finally the + sign for public. Under the Tables section, you should see a count of 167 in parentheses.
Configuring GenMAPP Builder to Connect to your PostgreSQL Database
- Launch gmbuilder.bat.
- If the program does not detect a database configuration, you will see a message window to this effect and the configuation dialog will open automatically once you close the message window. Otherwise:
- Select the menu item File > Configure Database...
- Under the Database Connections tab the Database Driver defaults to PostgreSQL. Enter information in the following fields:
- Host or address: localhost
- Port number: 5432
- Database name: VCholera_20151027_gmb3build5_KW
- Username: postgres
- Password: Welcome1
- Click the OK button.
Importing Data into the PostgreSQL Database
- Select File > Import UniProt XML...
- Navigate to the UniProt XML file that you extracted previously and click the Open button.
- This should take about 5-10 minutes, but may take longer depending on the size of the file, processor speed, and available memory of the machine. When the process has completed, record the elapsed time from the message window that appears.
- 2.96 min (had to redo this one)
- Select File > Import GO OBO-XML...
- Navigate to the GO OBO-XML file that you extracted previously. Click the Open button.
- This should take about 5-10 minutes, but may take longer depending on the size of the file, processor speed, and available memory of the machine. When the process has completed, record the elapsed time from the message window that appears.
- 6.88 min
Click OK to the message asking you to process the GO data.
- This should take about 5-10 minutes, but may take longer depending on the size of the file, processor speed, and available memory of the machine. When the process has completed, record the elapsed time from the message window that appears.
- 4.49 min
- Select File > Import GOA...
- Navigate to the GOA file that you downloaded previously and click the Import button. This process should only take a minute or so.
- 0.06 min
Exporting a GenMAPP Gene Database (.gdb)
- Select File > Export to GenMAPP Gene Database...
- Type a name in the Owner field (or else it won't let you export).
- When doing the individual exercise for the Week 9 assignment, use your own name. When doing this for your team project, use your team's name.
- GenMAPP Builder scans your PostgreSQL database to see what species are available. Click on the species that you would like to export, then click Next to continue.
- Create GenMAPP Database: click on the "Save GenMAPP Database File As..." button.
- In the Save dialog box that appears, navigate to the "T:" drive, and then modify the default file name by appending your initials. Click the "Save" button
- Leave the boxes checked for exporting all Molecular Function, Cellular Component, and Biological Process Gene Ontology Terms.
- Click the "Next" button to begin teh export process.
- Record the starting and ending times from the black console window. This will take 1-2 hours for a typical bacterial genome, depending on the size of the database, the processor speed, and available memory. Large eukaryotic genomes (like Arabidopsis thaliana) or genomes with many GO annotations (like Saccharomyces cerevisiae) can take much longer, in the range of 12-24 hours.
- NOTE: the progress bar is not accurate.
Checking the Quality of your Exported Gene Database
- It is a good idea to check the quality of your exported Gene Database to make sure that all of the data from the XML files made it into the PostgreSQL database and was then exported to the GenMAPP Gene Database. We have created a Gene Database Testing Report Sample to help guide you through this process.
TallyEngine
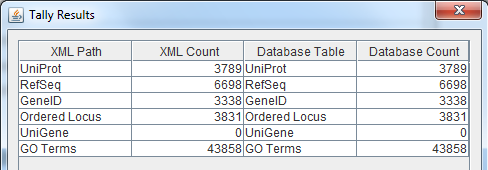
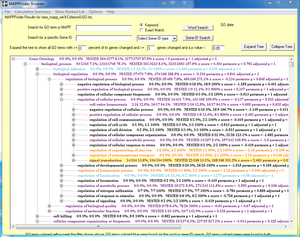
- Run the TallyEngine in GenMAPP Builder and record the number of records for UniProt and GO in the XML data and in the Postgres databases.
- Choose the menu item Tallies > Run XML and Database Tallies for UniProt and GO...
- Take a screenshot of the results. Upload the image to the wiki and display it on this page.
- For more information, see this page.
Using XMLPipeDB match to Validate the XML Results from the TallyEngine
Follow the instructions found on this page to run XMLPipeDB match.
- Are your results the same as you got for the TallyEngine? Why or why not?
- Using the command:
java - jar xmlpipedb-match-1.1.1.jar "VC_A?[0-9][0-9][0-9][0-9]" < uniprot-organism%3A243277.xml, the command line returnsTotal unique matches: 3831. These number of Gene ID's is the same as for the Tally Engine result.
- Using the command:
Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine
For more information, see this page.
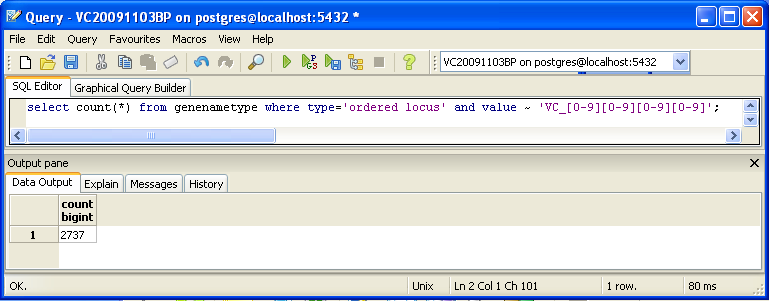
You can also look for counts at the SQL level, using some variation of a select count(*) query. This requires some knowledge of which table received what data. Here’s an initial tip: the gene/name tags in the XML file land in the genenametype table. A query on this table counting values from this table that were marked as ordered locus in the XML file matching the pattern VC_[0-9][0-9][0-9][0-9] would look like this:
select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_[0-9][0-9][0-9][0-9]';
In pgAdmin III, you can issue these queries by clicking on the pencil/SQL icon in the toolbar, typing the query into the SQL Editor tab, then clicking on the green triangular Play button to run.
- Are your results the same as reported by the TallyEngine? Why or why not?
- The above command returns 2737 gene ID's, because it does not account for ID's "VC_A####"
- However, using the command
select count(*) from genenametype where type = 'ordered locus' and value ~ 'VC_A?[0-9][0-9][0-9][0-9]'returns 3831 gene ID's, which is the same as for the XMLPipeDB Match and Tally Engine Results. This is because addingA>to the command allows pgAdminIII to include gene ID's that either do or don't include an "A" following the underscore.
OriginalRowCounts Comparison
Within the .gdb file, look at the OriginalRowCounts table to see if the database has the expected tables with the expected number of records. Compare the tables and records with a benchmark .gdb file.
Benchmark .gdb file: 2010 Benchmark Original Row Counts Table (found by Josh Kuroda on the 2010 course's wiki)
OriginalRowCounts table: media:KW_original_row_counts_from_gdbfile.xlsx (I copied this from Access and pasted it into an Excel file because a screenshot of this table would've been quite large).
The OrderedLocusNames table has the same amount of ID's (7664), however plenty of the values are very different, such as GeneOntology, which more than doubled between the benchmark and the new database.
Visual Inspection
Perform visual inspection of individual tables to see if there are any problems.
- Look at the Systems table. Is there a date in the Date field for all gene ID systems present in the database?
- No. 21 dates are missing.
- Open the UniProt, RefSeq, and OrderedLocusNames tables. Scroll down through the table. Do all of the IDs look like they take the correct form for that type of ID?
- UniProt - Yes. All appear to be of uniform format.
- OrderedLocusNames - No, some have underscores after "VA" and some do not.
- RefSeq - Yes, assuming different amounts of number characters is not considered to be a deviation from format.
Note:
.gdb Use in GenMAPP
Putting a gene on the MAPP using the GeneFinder window
This seems to work fine. All of the other ID's are present on the backpage.
Creating an Expression Dataset in the Expression Dataset Manager
- How many of the IDs were imported out of the total IDs in the microarray dataset? How many exceptions were there? Look in the EX.txt file and look at the error codes for the records that were not imported into the Expression Dataset. Do these represent IDs that were present in the UniProt XML, but were somehow not imported? Or were they not present in the UniProt XML?
- I used the Merrell et al. spreadsheet with my newly created .gdb file. Upon creating my expression dataset, I was presented with 121 errors. But this is actually the exact same number of errors reported when the 2010 database was used. This could mean one of two things: either the original GO-OBO-XML, GOA and UniProt files do not contain any significant updates (unlikely given a five-year gap, I think) or the Merrell et al. data itself is what's causing the errors.
- Upon opening the exceptions file, it can be seen that 5221 gene ID's were imported. However, this number includes the 121 errors.
- Adding a filter to column Y ("Errors") and opening the filter menu shows that the only error is "Gene not found in OrderedLocusNames or any related system." Further, applying a text filter for this error message returns 121 rows, verifying that this error message applies to all of the errors.
- It's hard to believe that the UniProt XML file wouldn't see a single update over a five year period, especially when there were many just between 2009 and 2010. So I'm inclined to believe this is an import issue.
Coloring a MAPP with expression data
This seems to work just fine.
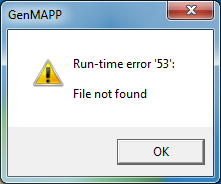
Running MAPPFinder
The MappFinder browser does generate the GO hierarchy, but clicking on any of the GO terms results in a run-time error. I'm not sure why.
Compare Gene Database to Outside Resource
The OrderedLocusNames IDs in the exported Gene Database are derived from the UniProt XML. It is a good idea to check your list of OrderedLocusNames IDs to see how complete it is using the original source of the data (the sequencing organization, the MOD, etc.) Because UniProt is a protein database, it does not reference any non-protein genome features such as genes that code for functional RNAs, centromeres, telomeres, etc.
Note:
Links
- Kevin Wyllie Week 2 (See the original assignment and class journal.)
- Kevin Wyllie Week 3 (See the original assignment and class journal.)
- Kevin Wyllie Week 4 (See the original assignment and class journal.)
- Kevin Wyllie Week 5 (See the original assignment and class journal.)
- Kevin Wyllie Week 6 (See the original assignment and class journal.)
- Kevin Wyllie Week 7 (See the original assignment and class journal.)
- Kevin Wyllie Week 8 (See the original assignment and class journal.)
- Kevin Wyllie Week 9 (See the original assignment and class journal.)
- Kevin Wyllie Week 10 (See the original assignment.)
- Kevin Wyllie Week 11 (See the original assignment.)
- Kevin Wyllie Week 12 (See the original assignment.)
- Kevin Wyllie Week 14 (See the original assignment.)
- Kevin Wyllie Week 15 (See the original assignment.)