GÉNialOMICS Gene Database Testing Report (Build 4 Export)
Contents
- 1 Export Information
- 2 Using TallyEngine
- 3 Using XMLPipeDB match to Validate the XML Results from the TallyEngine
- 4 Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine
- 5 OriginalRowCounts Comparison
- 6 Visual Inspection
- 7 .gdb Use in GenMAPP
- 8 Compare Gene Database to Outside Resource
Export Information
Version of GenMAPP Builder: GenMAPP Builder Custom, Build 4
Computer on which the export was run: Home Workstation
Postgres Database name: B.cenocepacia_J2315_20151204_BUILD4_genialomics
UniProt XML filename: uniprot-taxonomy%3A216591_GEN_BL12_20151119.xml
- UniProt XML version: UniProt release 2015_11 - November 11, 2015
- UniProt XML download link: UniProtKB link for the complete proteome of J2315
- Time taken to import: 3.46 minutes
- Note: Time taken appears to be slightly shorter than previous exports.
GO OBO-XML filename: go_daily-termdb_GEN_BL12_20151119.obo-xml
- GO OBO-XML version (derived from the date modified on the file, itself): Date Modified: 11/19/2015 2:24 AM
- GO OBO-XML download link: Link from GO website
- Time taken to import: 5.05 minutes
- Time taken to process: 3.75 minutes
- Note: Time taken appears to be slightly shorter than previous exports.
GOA filename: 31277.B_cepacia_GEN_BL12_20151119.goa
- GOA version: Date Modified: 11/10/15, 1:47:00 PM (information sourced from FTP site)
- GOA download link: FTP site file
- Time taken to import: 0.04 Minutes
- Note: No issues were found with the import of this file.
Name of .gdb file: Bc-Std GEN Build4 20151204.gdb
- Time taken to export: 11 hours 6 minutes
- Start time: 7:51 am
- End time: 6:57 pm
- Note: File was exported without any major issues, however, the export appeared to take significantly longer than the previous exports. It is likely that the export took so long because the workstation had, for some period of time, entered a "sleep" mode (export was delayed, as the computer had to be taken off of "sleep").
Using TallyEngine
- PostgreSQL was initialized through pgAdmin III and the database B.cenocepacia_J2315_20151204_BUILD4_genialomics was left running
- GenMAPP builder was booted and Run XML and Database Tallies for UniProt and GO was selected under the Tallies menu item; the UniProt XML and GO files that were imported were chosen
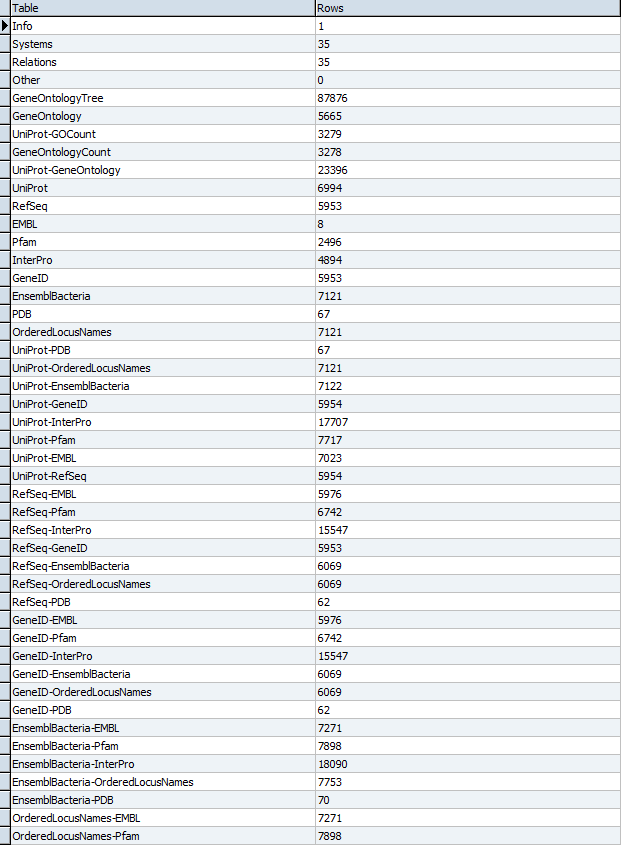
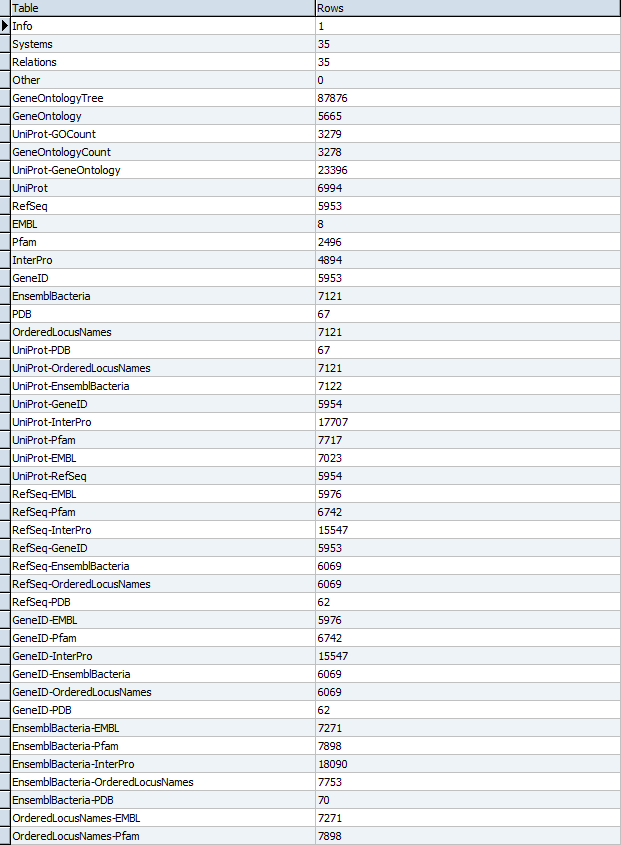
- Results of TallyEngine:
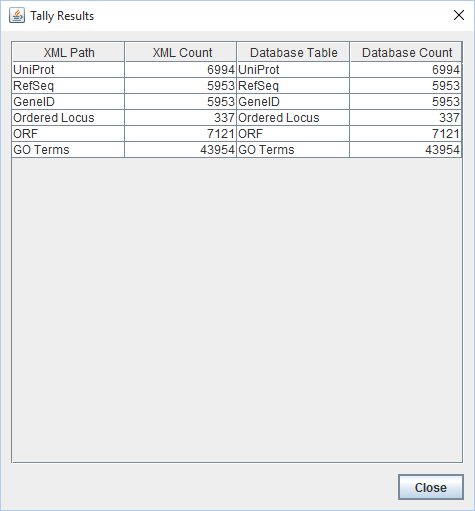
- Note: These results differ significantly from what was found in previous exports. The 337 Ordered Locus gene names are now distinct from the 7121 ORF gene names (and are represented, as such, by TallyEngine). All of the counts related to external references (like UniProt) remain the same. The major and crucial change is the inclusion and representation of the ORF data.
Using XMLPipeDB match to Validate the XML Results from the TallyEngine
- The Windows command line was launched (cmd.exe)
- This set of commands was inputted into the command line in order to utilize XMLPipeDB match to verify the OrderedLocusNames count:
java -jar xmlpipedb-match-1.1.1.jar "p?BCA[L,S,M]?[0-9][0-9][0-9][A,a]?[0-9]?[A-Z, a-z]?" < "uniprot-taxonomy%3A216591_GEN_BL12_20151119.xml"- NOTE: Prior to executing the command, the folder that held the files and xmlpipedb-match-1.1.1.jar was entered through the Windows command line (a set of CD commands was used in order to enter the correct directory). The results were identical to what was found in the the build 2 export.
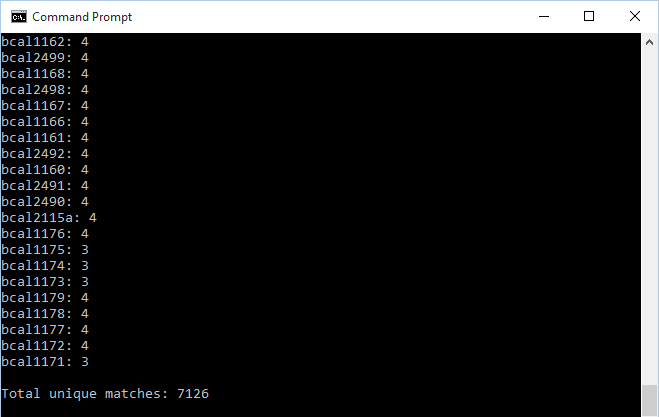
- 7126 unique matches were found through XMLPipeDB match
Are your results the same as you got for the TallyEngine? Why or why not?
- These results vary slightly from what was found by TallyEngine due to the presence of 5 discrepant IDs (which were identified in the Week 14 assignment). Barring those 5 IDs, the results by XMLPipeDB Match line up with what TallyEngine reports (since the Match query grabs ORF data).
Using SQL Queries to Validate the PostgreSQL Database Results from the TallyEngine
- pgAdmin III was booted and all of the necessary connections were made
- In pgAdmin III, the query
select count(*) from genenametype where type = 'ordered locus' and value ~ 'BceJ2315_[0-9][0-9][0-9][0-9]';was issued via the SQL Query menu in order to validate the TallyEngine count for "Ordered Locus" for the PSQL database.- 337 unique matches were found in pgAdmin III (postgres database results). This lines up with what was found in TallyEngine.
- Additionally, the query
select count(*) from genenametype where type = 'ORF' and value ~ 'p?BCA[A-Z]?[0-9][0-9][0-9][A-Z]?[0-9]?[A-Z, a-z]?';was run via SQL in order to the verify the ORF counts- 7121 counts were found which is identical to what was found through XMLPipeDB match (ignoring the discrepant IDs) and to what was reported by TallyEngine (for the ORF data).
- Are your results the same as reported by the TallyEngine? Why or why not?
- The results are now the same as what was reported by TallyEngine; this is due to the fact that the most recent build incorporated code fixes that allowed GenMAPP builder, and TallyEngine, to properly include the ORF data in their analysis/work.
OriginalRowCounts Comparison
- The newly created J2315 .gdb file (Bc-Std_GEN_BL12_20151201.gdb) was opened with a program that is able to explore a .mdb file (such as Microsoft Access); in this case, MDB Viewer Plus was utilized.
- Using the program, the OriginalRowCounts table was looked at, which contained summaries regarding each of the tables within the database (and the # of rows/entries in each of the tables)
- OriginalRowCounts for Build 4 export of J2315
- It was decided that a good reference or "benchmark" would be the database that was created using Build 3 of the customized GenMAPP builder; comparing the two should allow me to see if there was any difference in the imported data (and any difference in the functionality of GenMAPP builder).
- Benchmark .gdb file: compressed Bc-Std_GEN_Build3_20151203.gdb
- OriginalRowCounts for the Build 3 export of J2315
Note: It was noticed that the OriginalRowCounts table in this export are mostly identical to the one found through the Build 2 export. However, it was noticed that there existed differences in the OrderedLocusNames table between the two exports. It was found that the recent export, the Build 3 export, contained 7121 rows in the OrderedLocusNames table (which indicates 7121 entries, which is the same as the number of ORF gene names in the XML), while the last export, the Build 2 export, contained 337 rows in the OrderedLocusNames tables. The fact that the build 3 export how shows 7121 entries in that table is indicative of the fact that this modified GenMAPP builder (build 3) is now focusing on the ORF data; it appears, however, that it is now labeling the "ORF" data as being OrderedLocusNames instead of the "ordered locus" data. The observation in the OriginalRowCounts table does not completely mesh with what was found earlier in the PSQL database. In the PSQL database, it was found that the OrderedLocusName data was still the "ordered locus" gene names that reside in the XML (and the "ORF" data are the 7121 gene names of interest). In conclusion, it feels that there are some issues with TallyEngine and GenMAPP builder that are leading to some issues (such as TallyEngine not reporting the ORF data).
Visual Inspection
Perform visual inspection of individual tables to see if there are any problems.
- Look at the Systems table. Is there a date in the Date field for all gene ID systems present in the database?
- Yes, there are dates present for GeneOntology, InterPro, GeneID, RefSeq, UniProt, EMBL, PDB, Pfam, OrderedLocusNames, and EnsemblBacteria.
- Open the UniProt, RefSeq, and OrderedLocusNames tables. Scroll down through the table. Do all of the IDs look like they take the correct form for that type of ID?
- In the UniProt table, like before, it is apparent that only gene names of the type "ordered locus" are represented (no signs of gene names that begin with something like "BCA"). The RefSeq table appears to not have any problems. The ordered locus names table, now, only reflects gene names in the form of
p?BCA[L,M,S]?[0-9][0-9][0-9][A,a]?[0-9]?[A-Z, a-z]; it appears that the "ORF" data replaced the "ordered locus" gene names in this table (these IDs appear to be in the correct and common form).
Note: The modifications to GenMAPP builder appear to have changed some of the data within the tables of the gene database (ORF gene names replacing "ordered locus" gene names, with respect to the OrderedLocusNames table).
.gdb Use in GenMAPP
Note: To do.
Compare Gene Database to Outside Resource
Outside Resource: Burkholderia Genome DB, UniProt KB
- The strain page for J2315 was looked up: [1]
- 7121 OrderedLocusNames were found within the exported gdb file. 6,994 entries corresponding to protein encoding genes were found in UniProt KB, and 7114 coding sequences were found in the [http://beta.burkholderia.com/strain/show/146 MOD. It is apparent that the count of 7121 (ORF data) is much closer to what is present in outside resources than the one of 337 ("ordered locus" data). The differences in count between UniProt and the gdb and MOD could be the result of the fact that UniProt only covers genes that code for protein (some of the coding sequences present in the MOD, or within the gdb, could be responsible for functional RNA, which are not covered by UniProt).
- Note: The exported database now seems more in-line with what is to be expected of the genome of B. cenocepacia; the current OrderedLocusName counts (which actually represents ORF counts) seem very close to the counts expressed by the MOD and by UniProt.
| Weekly Group Assignments | Shared Group Journals | Project Links | Team Members |
|---|---|---|---|
|
|
|
|
|
Brandon Litvak
BIOL 367, Fall 2015
| Weekly Assignments | Individual Journal Pages | Shared Journal Pages |
|---|---|---|
|
|
|
|