Kevin Wyllie Week 15
From LMU BioDB 2015
Electronic Lab Notebook
MAPPFinder Procedure
- MAPPFinder was launched from the GenMAPP window: "Tools" > "MAPPFinder".
- "Calculate New Results" was selected from the MAPPFinder window.
- "Find File" was selected and in the directory, the .gex file (created in the Expression Dataset Manager - see week 14) was selected.
- This file can be found here.
- Under the "Select Color Set" field, "KWVP_20151205" was selected (the color set's name in this case is the same as the .gex file's name - and there's only one color set in the .gex file).
- After the color set has been selected, under the "Select Criteria to filter by" field, either increase or decrease was chosen.
- The boxes for "Gene Ontology" and "Click here to calculate p values..." were checked.
- The location to save the resulting .txt file was selected with the "Browse" option toward the bottom of the window. Note that because this protocol was carried out both for increase and decrease, there were two text files generated in MAPPFinder.
- "Run MAPPFinder" was selected to generate the gene ontology tree (and the aforementioned .txt files).
- The files generated can be found here:
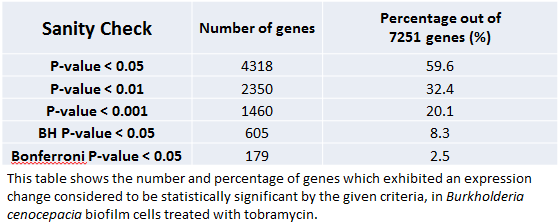
Sanity Check
Links
| Weekly Group Assignments | Shared Group Journals | Project Links | Team Members |
|---|---|---|---|
|
|
|
|
|
- Kevin Wyllie Week 2 (See the original assignment and class journal.)
- Kevin Wyllie Week 3 (See the original assignment and class journal.)
- Kevin Wyllie Week 4 (See the original assignment and class journal.)
- Kevin Wyllie Week 5 (See the original assignment and class journal.)
- Kevin Wyllie Week 6 (See the original assignment and class journal.)
- Kevin Wyllie Week 7 (See the original assignment and class journal.)
- Kevin Wyllie Week 8 (See the original assignment and class journal.)
- Kevin Wyllie Week 9 (See the original assignment and class journal.)
- Kevin Wyllie Week 10 (See the original assignment.)
- Kevin Wyllie Week 11 (See the original assignment.)
- Kevin Wyllie Week 12 (See the original assignment.)
- Kevin Wyllie Week 14 (See the original assignment.)
- Kevin Wyllie Week 15 (See the original assignment.)