Difference between revisions of "Dbashour Week 12"
From LMU BioDB 2017
(→List of 10 Unknown Words and Their Definitions: finished definitions) |
(→Cold shock and microarray procedures of yeast samples: syntax) |
||
| (4 intermediate revisions by the same user not shown) | |||
| Line 13: | Line 13: | ||
# midlgarithmic - cell numbers increase in a logarithmic fashion but have not reached their full reproduction rate (Rogers & Kadner, 2017) | # midlgarithmic - cell numbers increase in a logarithmic fashion but have not reached their full reproduction rate (Rogers & Kadner, 2017) | ||
# 2-fold - method of reporting statistics using log2 that results in more reproducible gene lists than do the ordinary and modified t-statistics (Witten & Tibshirani, 2007) | # 2-fold - method of reporting statistics using log2 that results in more reproducible gene lists than do the ordinary and modified t-statistics (Witten & Tibshirani, 2007) | ||
| + | |||
| + | = Flow Chart of Overall Experimental Design = | ||
| + | |||
| + | [[File: Flowchart_Design.png]] | ||
=Outline of Article= | =Outline of Article= | ||
| Line 22: | Line 26: | ||
* '''The purpose of this experiment is to analyze global gene expression in low temperature-exposed yeast cells using a yeast cDNA microarray to obtain fundamental information on low temperature response and low temperature-dependent gene expression in yeast cells''' | * '''The purpose of this experiment is to analyze global gene expression in low temperature-exposed yeast cells using a yeast cDNA microarray to obtain fundamental information on low temperature response and low temperature-dependent gene expression in yeast cells''' | ||
== Cold shock and microarray procedures of yeast samples == | == Cold shock and microarray procedures of yeast samples == | ||
| − | * S. cerevisiae YPH500 was used for all the analyses | + | * ''S. cerevisiae'' YPH500 was used for all the analyses |
* Cultured aerobically in YPD medium (yeast extract, peptone, and glucose) at 30°C and shaken at 100 rpm | * Cultured aerobically in YPD medium (yeast extract, peptone, and glucose) at 30°C and shaken at 100 rpm | ||
* 50 ml of the culture were collected for a reference time of 0 | * 50 ml of the culture were collected for a reference time of 0 | ||
| Line 41: | Line 45: | ||
* Fluorescence intensities were normalized | * Fluorescence intensities were normalized | ||
* Data was clustered and referred to the Munich Information Center for Protein Sequences functional database | * Data was clustered and referred to the Munich Information Center for Protein Sequences functional database | ||
| − | * Functional relationships among the genes in each cluster was determined | + | * Functional relationships among the genes in each cluster was determined |
| + | |||
== Results and Discussion == | == Results and Discussion == | ||
=== Global Expression Analysis of Low Temperature Response in Yeast Cells Using a cDNA Microarray === | === Global Expression Analysis of Low Temperature Response in Yeast Cells Using a cDNA Microarray === | ||
| Line 127: | Line 132: | ||
* Other organisms show similar responses in gene expression when exposed to environmental stresses | * Other organisms show similar responses in gene expression when exposed to environmental stresses | ||
| − | + | =Deliverable= | |
[[Media:Cold_Shock_Yeast_Genome_Response.pdf | Journal Club Week 12 Presentation]] | [[Media:Cold_Shock_Yeast_Genome_Response.pdf | Journal Club Week 12 Presentation]] | ||
| − | == | + | =Acknowledgements= |
| + | I met with Corrine outside of class to discuss the method of presenting for the journal club. We worked on our presentation together and equally prepared for the assignment. | ||
| + | While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source. <br> | ||
| + | [[User:Dbashour|Dbashour]] ([[User talk:Dbashour|talk]]) 11:57, 21 November 2017 (PST) | ||
| + | |||
| + | =References= | ||
| + | *AccessScience. (2015). Diauxic growth (diauxie). Retrieved November, 2017, from https://www.accessscience.com/content/diauxic-growth-diauxie/BR0105151 <br> | ||
| + | *Honee, V. (2009). De novo. Retrieved November, 2017, from http://www.biology-online.org/dictionary/De_novo <br> | ||
| + | *King, R.C., Mulligan, P.K., & Stansfield, W.D. (Eds.). (2014). A Dictionary of Genetics. In Oxford Reference. Retrieved from http://www.oxfordreference.com/view/10.1093/acref/9780199766444.001.0001/acref-9780199766444<br> | ||
| + | *Lackie, J. M. (2013). The dictionary of cell and molecular biology. Oxford: Academic Press. <br> | ||
| + | *LMU BioDB 2017. (2017). Week 12. Retrieved November 14, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_12 | ||
| + | *Lundin, C. (2005). Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Research, 33(12), 3799-3811. doi:10.1093/nar/gki681 <br> | ||
| + | * National Center for Biotechnology Information (2016). Trehalose. Retrieved November 2017, from https://pubchem.ncbi.nlm.nih.gov/compound/trehalose#section=Top <br> | ||
| + | *Ratini, M. (2016). Hypoxia and Hypoxemia. Retrieved November, 2017, from https://www.webmd.com/asthma/guide/hypoxia-hypoxemia#1 <br> | ||
| + | *Rogers, K., & Kadner, R. J. (2017). Growth of bacterial populations. Retrieved November, 2017, from https://www.britannica.com/science/bacteria/Growth-of-bacterial-populations#ref955458 <br> | ||
| + | *Sahara, T., Goda, T., & Ohgiya, S. (2002). Comprehensive expression analysis of time-dependent genetic responses in yeast cells to low temperature. Journal of Biological Chemistry, 277(51), 50015-50021. | ||
| + | *Shaw, P. E. (2002). Peptidyl‐prolyl isomerases: A new twist to transcription. EMBO reports, 3(6), 521-526. <br> | ||
| + | *Witten, D., & Tibshirani, R. (2007). A comparison of fold-change and the t-statistic for microarray data analysis. Analysis, 1776, 58-85. <br> | ||
| − | + | {{template:dbashour}} | |
Latest revision as of 22:13, 21 November 2017
Contents
- 1 Article
- 2 List of 10 Unknown Words and Their Definitions
- 3 Flow Chart of Overall Experimental Design
- 4 Outline of Article
- 4.1 Background Information on Purpose of Experiment
- 4.2 Cold shock and microarray procedures of yeast samples
- 4.3 Results and Discussion
- 4.3.1 Global Expression Analysis of Low Temperature Response in Yeast Cells Using a cDNA Microarray
- 4.3.2 Clustering Analysis of Global Expression Data
- 4.3.3 Genes related to transcription
- 4.3.4 Ribosomal Protein Genes
- 4.3.5 Cell Rescue, Defense, Death, and Aging
- 4.3.6 Metabolism and Energy Production
- 4.3.7 Signal Transduction Components
- 4.4 Conclusion
- 5 Deliverable
- 6 Acknowledgements
- 7 References
Article
Sahara, T., Goda, T., & Ohgiya, S. (2002). Comprehensive expression analysis of time-dependent genetic responses in yeast cells to low temperature. Journal of Biological Chemistry, 277(51), 50015-50021.
List of 10 Unknown Words and Their Definitions
- hypoxia - low oxygen levels in the blood (Ratini, 2016)
- ubiquitin - plays a role in the heat-shock response, is involved in quality control of nascent proteins, membrane trafficking, cell signalling, cell cycle control, X chromosome inactivation and the maintenance of chromosome structure (Lackie, 2013)
- Cytosolic - contents of the fluid in the cytoplasm of a cell (King, et al., 2014).
- diauxic shift - the two-phase growth response seen in a culture of microorganisms making a phenotypic adaptation to the addition of a second substrate (AccessScience, 2015)
- methyl methanesulfonate - A DNA damaging agent to induce mutagenesis and in recombination experiments (Lundin, 2005).
- peptidyl-prolyl cis/trans- isomerases - catalyse the cis–trans isomerisation of peptide bonds N-terminal to proline residues in polypeptide chains, play a role in the folding of newly synthesised proteins, and assist in cell cycle control (Shaw, 2002)
- trehalose - a sugar thought to be implicated in anhydrobiosis - the ability of plants and animals to withstand prolonged periods of desiccation (National Center for Biotechnology Information, 2016)
- de novo - New; not present previously; just beginning (Honee, 2009)
- midlgarithmic - cell numbers increase in a logarithmic fashion but have not reached their full reproduction rate (Rogers & Kadner, 2017)
- 2-fold - method of reporting statistics using log2 that results in more reproducible gene lists than do the ordinary and modified t-statistics (Witten & Tibshirani, 2007)
Flow Chart of Overall Experimental Design
Outline of Article
Background Information on Purpose of Experiment
- Low temperatures are known to have several effects on biochemical and physiological properties in various cells
- Cold shock proteins are induced when cells are exposed to low temperatures in order to cope with the drastic change in environment
- In yeast, the NSR1, TIP1, and OLE1 genes have been identified as important cold-inducible genes through past research
- low temperature-dependent gene expression and low temperature response are still unclear
- The purpose of this experiment is to analyze global gene expression in low temperature-exposed yeast cells using a yeast cDNA microarray to obtain fundamental information on low temperature response and low temperature-dependent gene expression in yeast cells
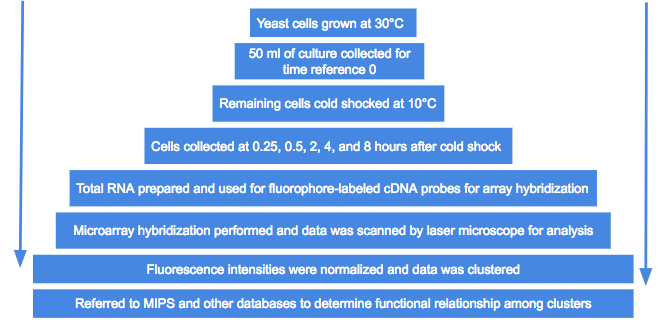
Cold shock and microarray procedures of yeast samples
- S. cerevisiae YPH500 was used for all the analyses
- Cultured aerobically in YPD medium (yeast extract, peptone, and glucose) at 30°C and shaken at 100 rpm
- 50 ml of the culture were collected for a reference time of 0
- Cells flash-frozen in liquid nitrogen
- Stored at -80°C in preparation for RNA
- The remaining cells were cold shocked at 10°C then cultured at the same temperature
- Cells collected at 0.25, 0.5, 2, 4, and 8 hours after the cold shock
- Cells flash-frozen in liquid nitrogen
- Stored at -80°C in preparation for RNA
- Cy3-dUTP and Cy5-dUTP were used as cDNA probes
- Labeled with fluorophore in order to carry out microarray hybridization
- Microarray hybridization performed based on the manual for S. cerevisiae cDNA microarray
- Microarrays were scanned by laser microscope
- Process repeated twice
- Averaged the expression ratios of the separate experiments for final data
- Images analyzed by computer program
- Data analyzed by analysis software
- Fluorescence intensities were normalized
- Data was clustered and referred to the Munich Information Center for Protein Sequences functional database
- Functional relationships among the genes in each cluster was determined
Results and Discussion
Global Expression Analysis of Low Temperature Response in Yeast Cells Using a cDNA Microarray
- Changes in expression of genes in yeast after cold shock was analyzed using cDNA of 5,803 genes in a yeast genome
- Ratio of fluorescent intensities was 2 fold for a mainly all the cDNA spots on the data
- Roughly 25% of yeast genes' expression levels were affected by cold shock
- Increase in number of genes up-regulated
- Increase in number of genes down-regulated
- Significantly up or down-regulated genes were classified according to the MIPS functional database
- Up-regulated and down-regulated genes increased in almost all categories when exposed to low temperatures
- Suggests that changes in gene expression are due to the introduction of low temperatures in order to adapt to their environment
- Other cells have demonstrated this ^^ when exposed to other environmental stresses
Clustering Analysis of Global Expression Data
- Genes close together were placed into clusters
- Clusters showed:
- Similar functions
- Cooperative regulation
- Clusters:
- 1A: Unclassified proteins
- 1B: Amino acid biosynthesis and metabolism
- 1C: RNA polym. I & RNA processing
- 1D: Ribosomal proteins
- 1E: Not defined
- Up regulated genes were divided into three clusters depending on their expression profiles
- IC: up-regulated after within 30 mins after cold shock
- ID: high up-regulation at 2hr and at 4-8 hr
- IE: high up-regulation at 2hr and at 4-8 hr
- Further classification of genes:
- 2A: RNA polymerase I & RNA processing - up regulated, then down regulated in later phase
- 2B: rRNA processing - up regulated, then down regulated in later phase
- 2C mRNA transcription - up-regulated in mid phase
- 2D: mRNA transcription - continuously down regulated
- These findings suggest that...
- The mechanisms for transcription are up regulated when exposed to low temperatures in the early phase
- Up-regulated mRNAs = essential for basic transcriptional/translational functions, like encoding regulatory proteins for amino acid production
- The genes for transcriptional regulation and mRNA synthesis made diverse responses in the late phase
- Down-regulated mRNAs = not essential for survival in cold shock, like genes encoding heat shock transcription factor or a transcription factor for drug resistant genes
- The mechanisms for transcription are up regulated when exposed to low temperatures in the early phase
Ribosomal Protein Genes
- Cluster Classification:
- 3A & 3B: cytosolic ribosome - up regulated first, then down regulated in later phase
- 3C: translational control factors - continuously up regulated
- 3D: tRNA synthetases - continuously down regulated
- These findings suggest that...
- Low temperature impairs translational ability
- Yeast genes up-regulate to compensate
Cell Rescue, Defense, Death, and Aging
- Cluster Classification:
- 4A: not labeled
- 4B & 4C: stress response - high up-regulation in mid-late phase
- 4D: stress response and chaperone - high down-regulation in mid-late phase
- These findings suggest that...
- Heat shock protein genes down-regulated (EXCEPT for HSP12 and HSP26)
- Protein folding genes up-regulated
Metabolism and Energy Production
- Cluster Classification:
- 5A: nucleotide metabolism - up-regulated early-mid phase, then down-regulated
- 5B & 5E: not specified
- 5C & 5D: C-compound and carbohydrate metabolism - continuously up-regulated
- 5F & 5H: amino acid metabolism - continuously down-regulated
- 5G: C-compound and carbohydrate utilization - continuously down-regulated
- These findings suggest that...
- Glycogen and trehalose production genes showed a lot of cooperative up-regulation
- Trehalose may help protect cellular membrane, which may help to keep yeast cells intact at low temperatures
Signal Transduction Components
- Cluster Classification:
- 6A: signal transduction - down-regulated
- 6B: signal transduction - up regulated in early phase, then down-regulation
- 6C: signal transduction - down-regulated in early phase, then up-regulation in late
- These findings suggest that...
- Genes related to cAMP-PKA pathway and Msn2p/4p were up-regulated
- Increase in PKA signaling has been known to be response to stresses
- Increase in...
- Signaling
- Metabolism control
- Stress resistance
Conclusion
- Gene expression changed to maintain transcription and translation and to adapt a tolerance to the colder temperature
- Transcriptional genes up-regulated first to help transcription and translation
- Ribosomal proteins up-regulated in the middle phase to further assist maintenance of translation
- Stress response induced genes up-regulated in the late phase
- Maintaining translation is priority to yeast in cold shock
- Cells need to make proteins to help maintain integrity and basic functions of cells
- Yeast cells can adapt to environment once ^^^ obtained to gain tolerance to low temperature
- Other organisms show similar responses in gene expression when exposed to environmental stresses
Deliverable
Journal Club Week 12 Presentation
Acknowledgements
I met with Corrine outside of class to discuss the method of presenting for the journal club. We worked on our presentation together and equally prepared for the assignment.
While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
Dbashour (talk) 11:57, 21 November 2017 (PST)
References
- AccessScience. (2015). Diauxic growth (diauxie). Retrieved November, 2017, from https://www.accessscience.com/content/diauxic-growth-diauxie/BR0105151
- Honee, V. (2009). De novo. Retrieved November, 2017, from http://www.biology-online.org/dictionary/De_novo
- King, R.C., Mulligan, P.K., & Stansfield, W.D. (Eds.). (2014). A Dictionary of Genetics. In Oxford Reference. Retrieved from http://www.oxfordreference.com/view/10.1093/acref/9780199766444.001.0001/acref-9780199766444
- Lackie, J. M. (2013). The dictionary of cell and molecular biology. Oxford: Academic Press.
- LMU BioDB 2017. (2017). Week 12. Retrieved November 14, 2017, from https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_12
- Lundin, C. (2005). Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Research, 33(12), 3799-3811. doi:10.1093/nar/gki681
- National Center for Biotechnology Information (2016). Trehalose. Retrieved November 2017, from https://pubchem.ncbi.nlm.nih.gov/compound/trehalose#section=Top
- Ratini, M. (2016). Hypoxia and Hypoxemia. Retrieved November, 2017, from https://www.webmd.com/asthma/guide/hypoxia-hypoxemia#1
- Rogers, K., & Kadner, R. J. (2017). Growth of bacterial populations. Retrieved November, 2017, from https://www.britannica.com/science/bacteria/Growth-of-bacterial-populations#ref955458
- Sahara, T., Goda, T., & Ohgiya, S. (2002). Comprehensive expression analysis of time-dependent genetic responses in yeast cells to low temperature. Journal of Biological Chemistry, 277(51), 50015-50021.
- Shaw, P. E. (2002). Peptidyl‐prolyl isomerases: A new twist to transcription. EMBO reports, 3(6), 521-526.
- Witten, D., & Tibshirani, R. (2007). A comparison of fold-change and the t-statistic for microarray data analysis. Analysis, 1776, 58-85.
List of Assignments
- Week 1
- Week 2
- Week 3
- Week 4
- Week 5
- Week 6
- Week 7
- Week 8
- Week 9
- Week 10
- Week 11
- Week 12
- Week 14
- Week 15
List of Individual Journal Entries
- dbashour Week 2
- dbashour Week 3
- dbashour Week 4
- dbashour Week 5
- dbashour Week 6
- dbashour Week 7
- dbashour Week 8
- dbashour Week 9
- dbashour Week 10
- dbashour Week 11
- dbashour Week 12
- dbashour Week 14
- dbashour Week 15
List of Shared Journal Entries
- Class Journal Week 1
- Class Journal Week 2
- Class Journal Week 3
- Class Journal Week 4
- Class Journal Week 5
- Class Journal Week 6
- Class Journal Week 7
- Class Journal Week 8
- Class Journal Week 9
- Class Journal Week 10
List of Final Assignments
List of Team Journal Assignments