Jnimmers Week 6
Jump to navigation
Jump to search
Biological Databases
Jnimmers
Assignment Table
Contents
- 1 Purpose
- 2 Questions
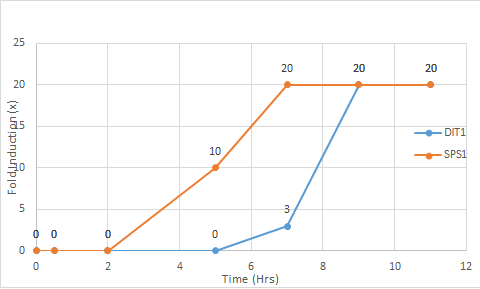
- 2.1 1. (Question 5, p. 110) Choose two genes from Figure 4.6b (PDF of figures on Brightspace) and draw a graph to represent the change in transcription over time. You can either create your plot in Excel and put the image up on your wiki page or you can do it by hand and upload a picture or scan.
- 2.2 2. (Question 6b, p. 110) Look at Figure 4.7, which depicts the loss of oxygen over time and the transcriptional response of three genes. These data are the ratios of transcription for genes X, Y, and Z during the depletion of oxygen. Using the color scale from Figure 4.6, determine the color for each ratio in Figure 4.7b. (Use the nomenclature "bright green", "medium green", "dim green", "black", "dim red", "medium red", or "bright red" for your answers.)
- 2.3 3. (Question 7, p. 110) Were any of the genes in Figure 4.7b transcribed similarly? If so, which ones were transcribed similarly to which ones?
- 2.4 4. (Question 9, p. 118) Why would most spots be yellow at the first time point? I.e., what is the technical reason that spots show up as yellow - where does the yellow color come from? And, what would be the biological reason that the experiment resulted in most spots being yellow?
- 2.5 5. (Question 10, p. 118) Go to the Saccharomyces Genome Database and search for the gene TEF4; you will see it is involved in translation. Look at the time point labeled OD 3.7 in Figure 4.12, and find the TEF4 spot. Over the course of this experiment, was TEF4 induced or repressed? Hypothesize why TEF4’s change in expression was part of the cell’s response to a reduction in available glucose (i.e., the only available food).
- 2.6 6. (Question, 11, p. 120) Why would TCA cycle genes be induced if the glucose supply is running out?
- 2.7 7. (Question 12, p. 120) What mechanism could the genome use to ensure genes for enzymes in a common pathway are induced or repressed simultaneously?
- 2.8 8. (Question 13, p. 121) Consider a microarray experiment where cells deleted for the repressor TUP1 were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later time points (glucose depleted) are labeled red. What color would you expect the spots that represented glucose-repressed genes to be in the later time points of this experiment?
- 2.9 9. (Question 14, p. 121) Consider a microarray experiment where cells that overexpress the transcription factor Yap1p were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented Yap1p target genes to be in the later time points of this experiment?
- 2.10 10. (Question 16, p. 121) Using the microarray data, how could you verify that you had truly deleted TUP1 or overexpressed YAP1 in the experiments described in questions 8 and 9?
- 3 Acknowledgements
- 4 References
Purpose
- The purpose of this assignment is learn about DNA assays, how they're carried out, their purpose and how to interpret the data collected from them.
Questions
1. (Question 5, p. 110) Choose two genes from Figure 4.6b (PDF of figures on Brightspace) and draw a graph to represent the change in transcription over time. You can either create your plot in Excel and put the image up on your wiki page or you can do it by hand and upload a picture or scan.
2. (Question 6b, p. 110) Look at Figure 4.7, which depicts the loss of oxygen over time and the transcriptional response of three genes. These data are the ratios of transcription for genes X, Y, and Z during the depletion of oxygen. Using the color scale from Figure 4.6, determine the color for each ratio in Figure 4.7b. (Use the nomenclature "bright green", "medium green", "dim green", "black", "dim red", "medium red", or "bright red" for your answers.)
- Gene X
- 1 Hr: Yellow
- 3 Hr:Bright Red
- 5 Hr: Black
- 9 Hr:Dim Green
- Gene Y
- 1 Hr: Yellow
- 3 Hr: Bright Red
- 5 Hr: Dim Green
- 9 Hr: Bright Green
- Gene Z
- 1 Hr: Yellow
- 3 Hr: Medium Red
- 5 Hr: Bright Red
- 9 Hr: Dim Red
3. (Question 7, p. 110) Were any of the genes in Figure 4.7b transcribed similarly? If so, which ones were transcribed similarly to which ones?
- Genes X and Y were both transcribed similarly. Both genes started at 1.0 as their ratio of transcription, in hour 3, hit their peak, and then declined from the 3rd hour to the 9th hour.
4. (Question 9, p. 118) Why would most spots be yellow at the first time point? I.e., what is the technical reason that spots show up as yellow - where does the yellow color come from? And, what would be the biological reason that the experiment resulted in most spots being yellow?
- The first time point contains yellow spots because the cells with glucose taken away are still operating with the glucose that they would have normally had, which is why the gene expression is the same. The overlapping green and red tags from the operation of both transcriptome transcribing the gene in what causes for the yellow spot because the two colors merge and produce the yellow marker.The yellow color indicates that the genes are both being transcribed in both the the transcriptomes, and so their working very similarly, so the yellow is the representation of that overlap.
5. (Question 10, p. 118) Go to the Saccharomyces Genome Database and search for the gene TEF4; you will see it is involved in translation. Look at the time point labeled OD 3.7 in Figure 4.12, and find the TEF4 spot. Over the course of this experiment, was TEF4 induced or repressed? Hypothesize why TEF4’s change in expression was part of the cell’s response to a reduction in available glucose (i.e., the only available food).
- Over the course of the experiment, TEF4 was repressed. This could be due to the cell not hang the necessary energy to express this gene due to the glucose levels of the cell being drastically reduced. Without the glucose, the cell won't have the ATP necessary to carry out certain functions, and so in an attempt to conserve energy, the cell cut out the expression of the TEF4 gene.
6. (Question, 11, p. 120) Why would TCA cycle genes be induced if the glucose supply is running out?
- The TCA cell can be induced as an option for the cell to somehow create ATP by producing molecules like NADH and FADH2. Through this process, the cell has options that can lead to the production of ATP, which it is in desperate need of with nearly no available glucose options. By keeping their metabolism going, even in this desperate way, the cell can stay alive by using any forms of acetyl CoA that they can scavenge without access to glycolysis.
7. (Question 12, p. 120) What mechanism could the genome use to ensure genes for enzymes in a common pathway are induced or repressed simultaneously?
- The genome could utilize negative and positive feedback loops in order to inhibit or promote the inducing or repressing of genes depending on a multitude of factors such as necessary nutrients, damages/mutations, and general necessity/production of the gene.
8. (Question 13, p. 121) Consider a microarray experiment where cells deleted for the repressor TUP1 were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later time points (glucose depleted) are labeled red. What color would you expect the spots that represented glucose-repressed genes to be in the later time points of this experiment?
- I would expect to see a red spot at a later time point because, similar to the earlier experiment, the glucose repression would lead to the suppression of the the gene that was being tested on, however, with TUP1 being deleted, there is no gene that would have the job of repressing the genes that are supposed to be repressed, meaning that you would see a green spot, indicating that (due to lack of repression) the cells are still expressing those genes in the later time points. However, I believe that the cell would be in much worse shape, metabolically, because it cannot shut off its gene expression.
9. (Question 14, p. 121) Consider a microarray experiment where cells that overexpress the transcription factor Yap1p were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented Yap1p target genes to be in the later time points of this experiment?
- The Yap1p target genes will likely show up as red due to an over expression of Yap1p means that the cell will likely be far more resistant to environmental stressors, allowing it to survive and freely express the Yap1p target genes even with low levels of glucose at later time points.
10. (Question 16, p. 121) Using the microarray data, how could you verify that you had truly deleted TUP1 or overexpressed YAP1 in the experiments described in questions 8 and 9?
- These genes can be verified as being deleted (TUP1) or over expressed(Yap1p) by looking at the color of the spots corresponding to the genes that each gene is targeting. For example, because TUP1 is a known repressor linked to glucose levels, you would expect for target gene expression to be suppressed when the levels of glucose are depleted. If a green spot isn't seen in this case, it can only be concluded that the operating systm used to repress that gene in this particular situation is not doing it's function(TUP1 in this case), so it is either mutated to the point of non function, or simply deleted. The same can be said for YAP1: if a target gene of YAP1 is still being expressed with a depleted glucose storage, either the repressor is deleted, or the gene that allows for resistance to glucose level change is working too well, which indicated the gene's over expression.
Acknowledgements
- Thank you to Dr. Kam Dahlquist for her assistance in this class and for helping to teach us about the topics covered in this assignment. Thank you for providing references necessary to answer these questions as well.
- Thank you to my homework partner, Yeabsira (Aby) Mesfin for his assitance on this assignment,. We met in person and spoke over the phone to coordinate questions and answers to the individual journal assignment questions.
- Except for what is noted above, this individual journal entry was completed by me and not copied from another source.
Jnimmers (talk) 19:38, 9 October 2019 (PDT)
References
- Alberts et al. (2002) Molecular Biology of the Cell, Ch. 8: Microarrays
- Brown, P.O. & Botstein, D. (1999) Exploring the new world of the genome with DNA microarrays Nature Genetics 21: 33-37.
- Campbell, A.M. and Heyer, L.J. (2003), “Chapter 4: Basic Research with DNA Microarrays”, in Discovering Genomics, Proteomics, and Bioinformatics, Cold Spring Harbor Laboratory Press, pp. 107-124.
- DeRisi, J.L., Iyer, V.R., and Brown, P.O. (1997) Exploring the Metabolic and Genetic Control of Gene Expression on a Genomic Scale. Science 278: 680-686.
- LMU BioDB 2019. (2019). Week 6. Retrieved October 10, 2019, from https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/Week_6