Marmas Week 6
Contents
Purpose
The purpose of this experiment is to develop an understanding of DNA microarray experiments and their function by answering questions concerning their analysis. Additionally, a case study will be looked at that examines a violation of the standards of research ethics.
Questions
Question 1
Question 2
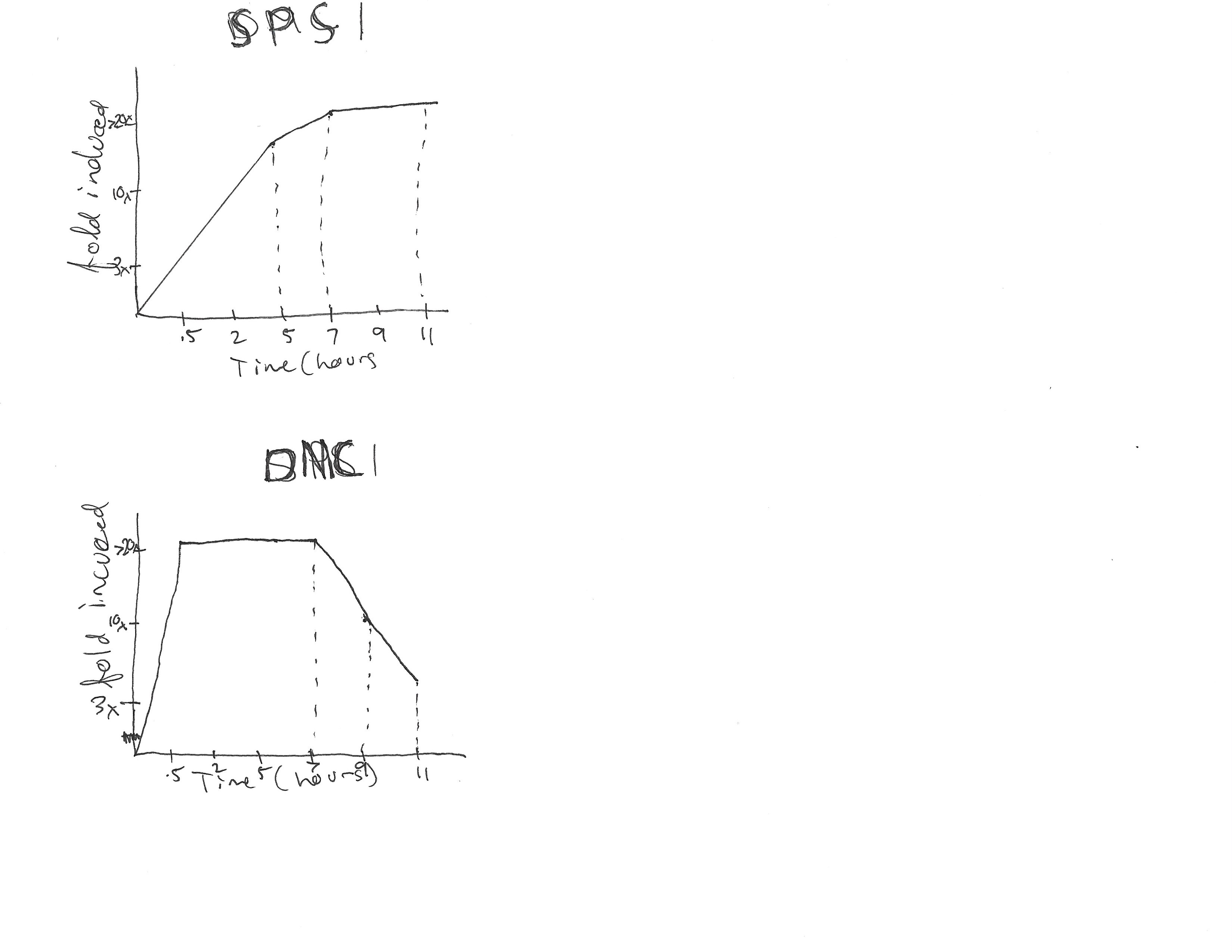
Transcriptional Response of Three genes to the gradual loss of oxygen
- Gene X
- Hour 1: Black
- Hour 3: Medium Red
- Hour 5: Black
- Hour 9: Bright Green
- Gene Y
- Hour 1: Black
- Hour 3: Bright Red
- Hour 5: Dim Green
- Hour 9: Bright Green
- Gene Z
- Hour 1: Black
- Hour 3: Dim Red
- Hour 5: Medium Red
- Hour 9: Medium Red
Question 3
Genes X and Y were transcribed very similarly, except for at hour 3. However, looking at the ratio of activity, they are very similar at each hour.
Question 4
On a color spectrum, yellow is in between red and green. Thus, the color yellow, which on microarrays represents no change, shows that the gene is neither being induced or repressed. As it becomes more repressed, the color will shift to a shade of green. As it becomes more induced, it will shift to a shade of red. Looking at a microarray, there are some genes that show orange or a yellow-green color. This is an example of a less potent induction or repression as compared to a strong induction or strong repression. In an experiment, the gene expression may not change, thus showing a yellow spot.
Question 5
As the experiment progressed the expression of TEF4 was repressed. This is indicative of the yellow-green spot on the microarray at OD 3.7. An increase in OD is indicative of more yeast cells being present in the solution. This means that the glucose, which is the only available food source in the system, is being rapidly consumed by the increasing number of yeast cells. With a decrease in available glucose, the cell shifts away from the translation of proteins as it cannot afford to make more in a stressed condition. It may even be using proteins to survive in these conditions. Therefore, TEF4, which is involved in translation, is no longer needed for survival and its expression is repressed.
Question 6
As glucose levels decrease, the cell needs to survive by making its own glucose. To do this, genes associated with the TCA cycle are up regulated to make more glucose for the cell. In a stressed condition, this is an expected response for survivability.
Question 7
A genome can regulate the transcription of a gene by the use of transcription factors. As an enzyme is produced, it may may cause the repression of a different enzyme by serving to either up regulate or down regulate expression. Then, when one enzyme is less abundant, the second enzyme will be produced and can serve to repress the other enzyme. Often, transcription factors are modified so they are either performing more transcription (induction) or are unable to transcribe the gene (repression).
Question 8
In cells where the repressor gene TUP1 was deleted, the cell would not be able to repress the expression of genes in a stressed environment. Therefore, after t = 0, it would be expected that the expression of genes would not be regulated and that spots for many genes would turn red. However, after more time, it's expected for these spots to turn back to green, as the cell would die after a short period time and genes would no longer be expressed. The cell would not able to control how its resources are used and will continue to allocate them to where is not necessary for survival.
Question 9
Just as when a repression factor is turned off, the over expression of a transcription factor would induce the transcription of various genes for the cell. At the start time, the cells spots for many genes would be green, but would turn red over time because of the over induced transcription. As the experiment progresses, it would be expected for the cells to die, meaning the spots would once again turn green. This is due to the cell not being able to control where its resources are allocated.
Question 10
To determine if the TUP1 was deleted or if YAP1 was over expressed, the microarray data would show the many green spots at the starting time. Then, as the experiment progresses, an overwhelming amount of spots would show red, as they are being forced to be expressed more often than normal. After more time progressed, the spots would overwhelmingly shift back to green. At this point, the many cells would have died due to not being able to survive in stressful conditions. Additionally, it would help to have a control experiment for comparison. The time at which the cells died in the control group (as the would normally once glucose levels were exhausted) would be much greater than that of the experimental group. The time for the cells in the experimental group would be much shorter in the control group.
Acknowledgements
- I would like to acknowledge my homework partner, David Ramirez, for aiding in the understanding of the analysis questions.
- I would like to acknowledge Dr. Dahlquist for her continuous help in the understanding of this field.
Except for what is noted above, this individual journal entry was completed by me and not copied from another source.
Marmas (talk) 17:07, 9 October 2019 (PDT)
References
Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K., & Walter, P. (2002). Molecular biology of the cell. New York: Garland Science.
Brown, P. O., & Botstein, D. (1999). Exploring the new world of the genome with DNA microarrays. Nature genetics, 21(1s), 33.
Campbell, A.M. and Heyer, L.J. (2003), “Chapter 4: Basic Research with DNA Microarrays”, in Discovering Genomics, Proteomics, and Bioinformatics, Cold Spring Harbor Laboratory Press, pp. 107-124.
DeRisi, J.L., Iyer, V.R., and Brown, P.O. (1997) Exploring the Metabolic and Genetic Control of Gene Expression on a Genomic Scale. Science 278: 680-686.
LMU BioDB 2019. (2019). Week . Retrieved October 8th, 2019, from https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/Week_6