Msamdars Week 2
Contents
Electronic Lab Notebook
Purpose
The purpose of this experiment is to learn about models and modeling, as well as the central dogma of molecular biology by utilizing the software Aipotu. We will use this software to learn about the real computational work that systems biologists do and emulate that work with hypothetical organisms as a control.
Methods
Task A
- Loaded each allele in to Aipotu
- Created pure-breeding species by continuously self-breeding each initial flower in the Genetics tab.
- Created hybrids between each pure-breeding homozygous flower and a pure-breeding homozygous White flower
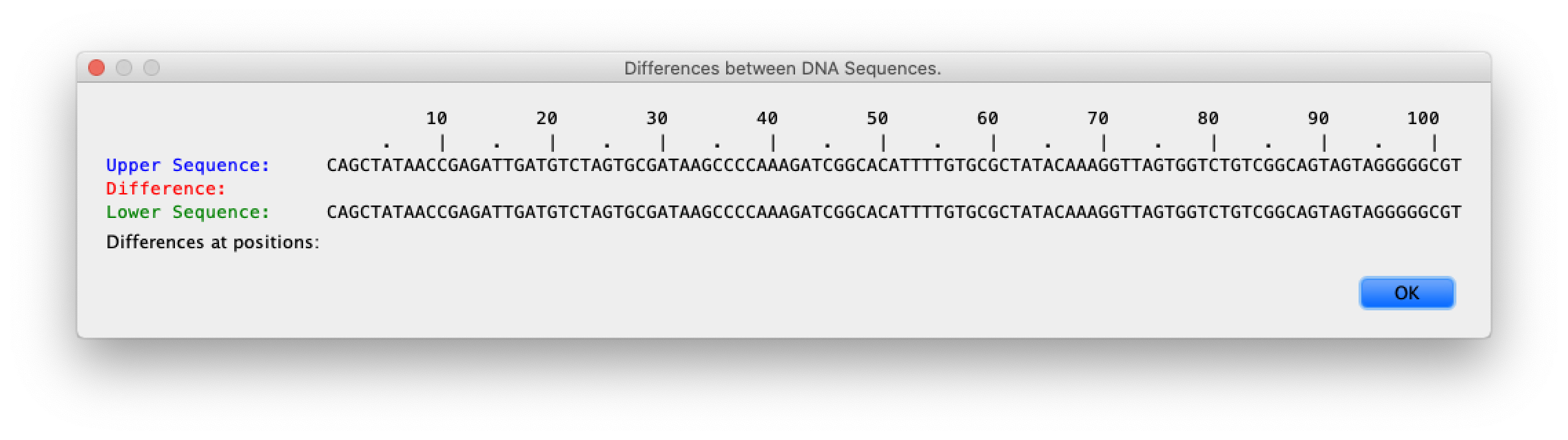
- Compared each of these hybrids's DNA seq by using built in compare tool, shown in a screenshot below.
- Compared each of the amino acid sequences by hand, copying the sequence over from the Molecular Bio screen into the table in the Results section.
Task B
- Loaded sample by double-clicking on White flower organism in Greenhouse (molecular biology tab in Aipotu pictured on right).
- Compared sequence using compare tool
Task C
- Loaded each sample into the Molecular Biology tab.
- Examined and recorded the color boxes in bottom right of each sample window for each organism.
Task D
- Created a cross of pure-breeding homozygous Blue flower and pure-breeding homozygous Red flower.
- Compared the proteins synthesized by the co-dominant alleles of the hybrid Purple flower.
- Determined the difference between the two proteins
- Created hypothesis that adding amino acids with aromatic groups (specifically Phenylalanine from the Blue allele) around locations of amino acids with aromatic groups would change color.
- This is also similar in location to the locations where most of the variation between the alleles happens.
- Tested hypothesis
- Recorded result
Task F
- Looked up single-letter codes for amino acids (John et al., 2017)
- Utilized Biochemistry window to type in the single letter codes
- The entry window automatically translates single letter inputs via the keyboard into the appropriate amino acids.
- Pressed Fold in the menu
- Recorded results
Results
Task A
*Differences in DNA or protein sequence from that of the white allele are underlined and red.
| Allele | Color | Change(s) in Amino Acid Sequence | Change(s) in DNA Sequence |
|---|---|---|---|
| B | Blue | CAGCTATAACCGAGATTGATGTCTAGTGCGATAAGCCCCAAAGATCGGCACATTTTGTGCGCTATACAAAGGTTAGTGTACTGTCGGCAGTAGTAGGGGGCGT
|
N-MetSerAsnArgHisIleLeuLeuValTyrCysArgGln-C
|
| R | Red | CAGCTATAACCGAGATTGATGTCTAGTGCGATAAGCCCCAAAGATCGGCACATTTTGTGCGCTATACAAAGGTTAGTGTTCTGTCGGCAGTAGTAGGGGGCGT
|
N-MetSerAsnArgHisIleLeuLeuValPheCysArgGln-C
|
| G | Green | CAGCTATAACCGAGATTGATGTCTAGTGCGATAAGCCCCAAAGATCGGCACATTTTGTGCGCTATACAAAGGTTAGTGTACTGGCGGCAGTAGTAGGGGGCGT
|
N-MetSerAsnArgHisIleLeuLeuValTyrTrpArgGln-C
|
| Y | Yellow | CAGCTATAACCGAGATTGATGTCTAGTGCGATAAGCCCCAAAGATCGGCACATTTTGTGCGCTATACAAAGGTTAGTGTGGTGTCGGCAGTAGTAGGGGGCGT
|
N-MetSerAsnArgHisIleLeuLeuValTrpCysArgGln-C
|
| W | White | CAGCTATAACCGAGATTGATGTCTAGTGCGATAAGCCCCAAAGATCGGCACATTTTGTGCGCTATACAAAGGTTAGTGGTCTGTCGGCAGTAGTAGGGGGCGT
|
N-MetSerAsnArgHisIleLeuLeuValValCysArgGln-C
|
Task B
Given the screenshot on the right, there were no differences in the white alleles.
Task C
The DNA sequences present in each of the starting organisms are given in the table above. The table below describes the allele composition of each of the four starting organisms.
| Organism Name | Allele 1 | Allele 2 |
|---|---|---|
| White | White | White |
| Green-1 | Green | Green |
| Green-2 | Blue | Yellow |
| Red | Red | White |
Task D
A purple flower is created by adding Phenylalanine codon to the DNA that encodes the Red allele, or a Tyrosine codon to the DNA that encodes the Blue protein. An example with the DNA fragment that encodes the Red allele is shown below, with the position of the codon underlined and in red. The amino acid sequence is shown below as well, with the position of the addition of Phenylalanine underlined and in red. Because the DNA sequence encodes for a protein that is itself purple, it doesn't rely on the co-dominance of blue and red alleles in order to make a purple-colored protein, thus it will be pure-breeding and purple.
DNA Sequence
CAGCTATAACCGAGATTGATGTCTAGTGCGATAAGCCCCAAAGATCGGCACATTTTGTGCGCTATACAAAGGTTAGTGTATTTCTGTCGGCAGTAGTAGGGGGCGT
Amino Acid Sequence
N-MetSerAsnArgHisIleLeuLeuValTyrPheCysArgGln-C
Task E
Because of the central model of molecular biology, a single mutation in the DNA sequence changes the corresponding mRNA, which in turn changes the composition of the amino acid sequence of the proteins. Thus, the variation in the alleles in combination with the mutations creates the variation in the effects that we saw in Part I.
Task F
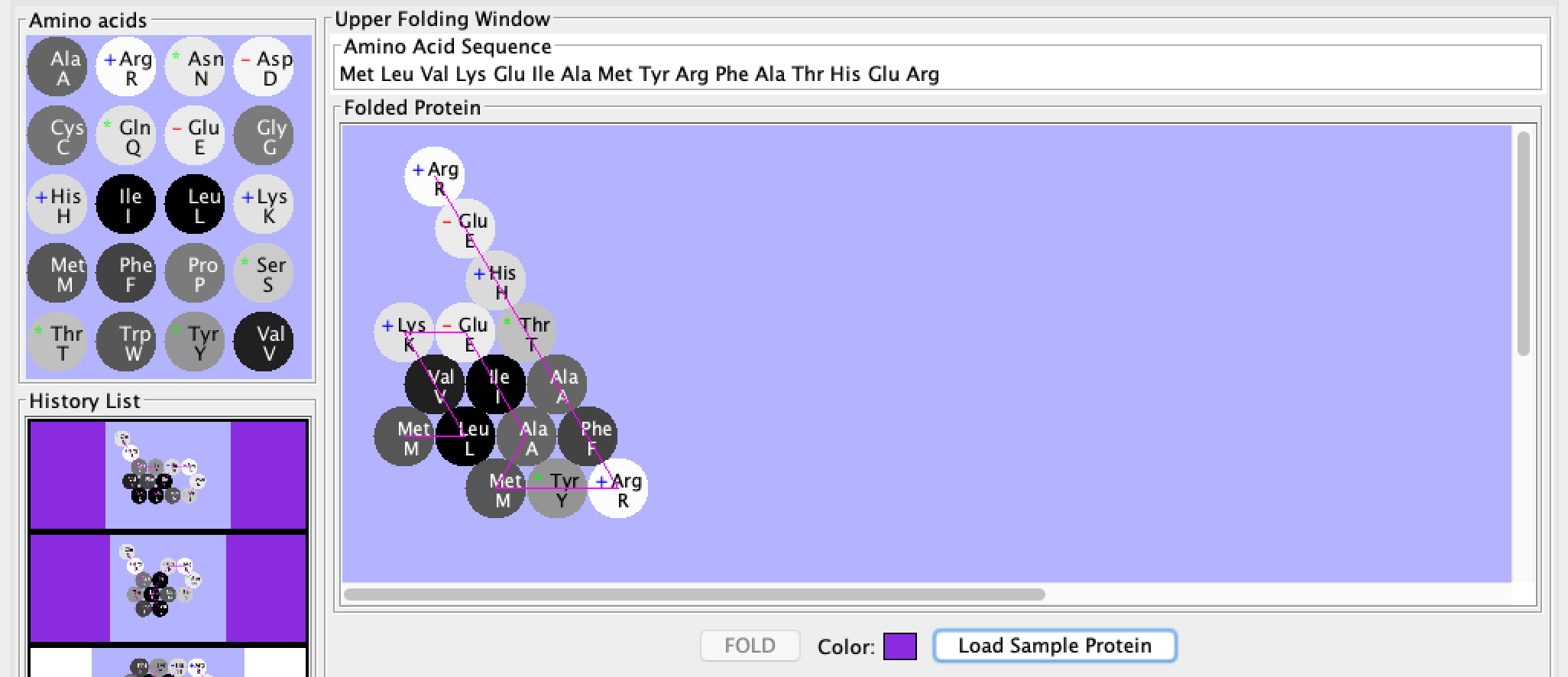
The results of creating the protein specified by the assignment are shown in the screenshot to the right. The single letter codes and their corresponding amino acids are shown in the left box, the amino acid sequence in the top box, the folded protein in the large box, and the final color on the bottom.
Conclusion
During the course of this project, I better understood how the variations in genetic information can have an impact on the downstream mRNA and amino acid sequences. Furthermore, by doing this project, I understood how scientists use more complicated tools to analyze organismal DNA and amino acid compositions in organisms that are not the sample organisms provided by the Aipotu. We identified how insertions and mutations can change amino acid sequences and the function of the proteins. I identified how tiny changes in the alleles combined with other tiny changes in alleles can create entirely new phenotypes.
Other Links
| Links | ||
|---|---|---|
| Mihir Samdarshi User Page | ||
| Assignment Pages | Personal Journal Entries | Shared Journal Entries |
| Week 1 | Journal Week 1 | Class Journal Week 1 |
| Week 2 | Journal Week 2 | Class Journal Week 2 |
| Week 3 | FAS2/YPL231W Week 3 | Class Journal Week 3 |
| Week 4 | Journal Week 4 | Class Journal Week 4 |
| Week 5 | Database - AmtDB | Class Journal Week 5 |
| Week 6 | Journal Week 6 | Class Journal Week 6 |
| Week 7 | Journal Week 7 | Class Journal Week 7 |
| Week 8 | Journal Week 8 | Class Journal Week 8 |
| Week 9 | Journal Week 9 | Class Journal Week 9 |
| Assignment Pages | Personal Journal Entries | |
| Week 10 | Journal Week 10 | |
| Week 11 | Journal Week 11 | |
| Week 12/13 | Journal Week 12/13 | |
| Team Project Links | ||
| Skinny Genes Team Page | ||
Acknowledgements
This week my partner was John Nimmers-Minor. We worked in and out of class together to better understand the questions. I also asked Mike Armas for assistance with Task D, as he was working on the Biochemistry portion of this assignment.
Except for what is noted above, this individual journal entry was completed by me and not copied from another source.
References
- LMU BioDB 2019. (2019). Week 2. Retrieved September 11, 2019, from https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/Week_
- MediaWiki (2019). Category: Help. Retrieved September 11, 2019, from https://www.mediawiki.org/wiki/Category:Help
- John, G.S., Rose, C., & Takeuchi, S. (2017). 9 Understanding Tools and Techniques in Protein Structure Prediction.