Difference between revisions of "Hivanson Week 10"
(→Clustering and GO Term Enrichment with stem: left off ## I clicked the "Browse..." button to the right of the "Gene Annotation File" item. Browse to your "stem" folder and select the file "gene_association.sgd.gz" and click Open.) |
(→Conclusion: conclusion complete) |
||
| (55 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
===Purpose=== | ===Purpose=== | ||
| + | We are analyzing a DNA microarray dataset, involving clustering and analysis of gene regulatory networks using GRNsight. The DNA microarray dataset is of the ''S. cerevisiae'' genome's cold shock response in a ∆CIN5 strain. This analysis will enable us to understand the role of the transcription factor CIN5's role in the cold shock response in ''S. cerevisiae''. | ||
| + | |||
===Methods/Results=== | ===Methods/Results=== | ||
==== Clustering and GO Term Enrichment with stem ==== | ==== Clustering and GO Term Enrichment with stem ==== | ||
| Line 14: | Line 16: | ||
#** I saved the dCIN5_stem sheet as Text (Tab-delimited) (*.txt). | #** I saved the dCIN5_stem sheet as Text (Tab-delimited) (*.txt). | ||
# I downloaded and extracted the STEM software. [http://www.cs.cmu.edu/~jernst/stem/ Click here to go to the STEM web site]. | # I downloaded and extracted the STEM software. [http://www.cs.cmu.edu/~jernst/stem/ Click here to go to the STEM web site]. | ||
| − | # | + | #* I downloaded the Gene Ontology and yeast GO annotations from the [https://xmlpipedb.cs.lmu.edu/biodb/spring2024/index.php/Week_10|Week 10 Wiki page]. |
#* I launched the STEM program. | #* I launched the STEM program. | ||
# '''Running STEM''' | # '''Running STEM''' | ||
| Line 21: | Line 23: | ||
##* I checked the box next to ''Spot IDs included in the data file''. | ##* I checked the box next to ''Spot IDs included in the data file''. | ||
## In section 2 (Gene Info) of the main STEM interface window, I left the default selection for the three drop-down menu selections for Gene Annotation Source, Cross Reference Source, and Gene Location Source as "User provided". | ## In section 2 (Gene Info) of the main STEM interface window, I left the default selection for the three drop-down menu selections for Gene Annotation Source, Cross Reference Source, and Gene Location Source as "User provided". | ||
| − | ## I clicked the "Browse..." button to the right of the "Gene Annotation File" item. | + | ## I clicked the "Browse..." button to the right of the "Gene Annotation File" item. I selected the file "gene_association.sgd.gz" and click Open. |
| − | ## In section 3 (Options) of the main STEM interface window, | + | ## In section 3 (Options) of the main STEM interface window, I ensured that the Clustering Method says "STEM Clustering Method" and did not change the defaults for Maximum Number of Model Profiles or Maximum Unit Change in Model Profiles between Time Points. |
| − | ## In section 4 (Execute) | + | ## In section 4 (Execute) I clicked on the yellow Execute button to run STEM. |
| − | |||
# '''Viewing and Saving STEM Results''' | # '''Viewing and Saving STEM Results''' | ||
| − | ## A new window | + | ## A new window opened called "All STEM Profiles (1)". |
| − | ##* | + | ##* I clicked on the button that says "Interface Options...". At the bottom of the Interface Options window that appears below where it says "X-axis scale should be:", I clicked on the radio button that says "Based on real time". I then closed the Interface Options window. |
| − | ##* | + | ##*I screenshotted this window and pasted it into my [[Media:BIOL367_S24_hivanson_dCIN5_p-value_slide.pptx|PowerPoint presentation]]. |
| − | ## | + | ## I clicked on each of the significant/colored profiles to open a window showing a more detailed plot containing all of the genes in that profile. |
| − | ##* | + | ##* I screenshoted each of the individual profile windows and saved the images in my [[Media:BIOL367_S24_hivanson_dCIN5_p-value_slide.pptx|PowerPoint presentation]]. |
| − | ##* | + | ##* For each of the profiles, I clicked on the "Profile Gene Table" button, then clicked on the "Save Table" button to save the file for each profile. This can be found in the [[Hivanson_Week_10#Data_.26_Files|Data and Files]] section below. |
| − | + | ##* For each of the significant profiles, I clicked on the "Profile GO Table" to see the list of Gene Ontology terms belonging to the profile, then clicked on the "Save Table" button for each profile. This can be found in the [[Hivanson_Week_10#Data_.26_Files|Data and Files]] section below. | |
| − | ##* For each of the significant profiles, | ||
| − | |||
# '''Analyzing and Interpreting STEM Results''' | # '''Analyzing and Interpreting STEM Results''' | ||
| − | ## | + | ## I selected profile 22 for further intepretation of the data. |
| − | ##* | + | ##*'''Why did you select this profile? In other words, why was it interesting to you?''' |
| − | ##* | + | ##**I selected profile 22 because the overall trend of a spike only at 90 minutes is different from the rest of the profiles' trends. Further, it had the second highest number of genes belonging to it out of all significant profiles. |
| − | ##* | + | ##*'''How many genes belong to this profile?''' |
| − | ##* | + | ##**179 genes |
| − | ##* | + | ##*'''How many genes were expected to belong to this profile?''' |
| − | ##* | + | ##**22.9 genes |
| − | ##** | + | ##*'''What is the p value for the enrichment of genes in this profile?''' |
| − | + | ##**3.4E-98 | |
| − | ##**''''' | + | ##* I opened the GO list file that I saved for profile 22 in Excel. I selected the third row and then filtered on the "p-value" column to show only GO terms that have a p value of < 0.05. |
| − | ##** | + | ##* '''How many GO terms are associated with this profile at p < 0.05?''' |
| − | + | ##** 25 | |
| − | + | ##* I filtered on the "Corrected p-value" column to show only GO terms that have a corrected p value of < 0.05. | |
| − | + | ##* '''How many GO terms are associated with this profile with a corrected p value < 0.05?''' | |
| − | + | ##** 7 | |
| − | + | ##* I select 6 Gene Ontology terms from your filtered list | |
| − | + | ##** These were selected by choosing the six most significant terms that were not redundant with each other. | |
| − | + | ##**'''''I looked up the definitions for each of the terms at [http://geneontology.org http://geneontology.org].''''' | |
| − | + | ##**'''Why does the cell react to cold shock by changing the expression of genes associated with these GO terms?''' | |
| − | + | ##***The genes that are associated with these GO terms include stress responses, like <code>cellular response to oxidative stress</code>, and its related terms that I excluded from the above table due to similarity. This makes sense as cold shock is considered a stressor. Additionally, various cellular structure components are associated with these GO terms, such as <code>cytoplasm</code>, <code>cytoskeleton</code>, and <code>fungal-type vacuole</code> which all may need to have regulated rigidity, fluidity or some other physical characteristics at lower temperatures. | |
| − | + | ##**'''Also, what does this have to do with the transcription factor being deleted (for the groups working with deletion strain data)''' | |
| − | + | ##***I want to compare our results to that of the wildtype. The [https://www.yeastgenome.org/locus/S000005554 SGD description of CIN5] says it is <code>nuclearly localized under oxidative stress</code>, so perhaps with the deletion of CIN5 transcription factor, oxidative stress, and possibly other stress responses would be unable to occur. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
{| class="wikitable" | {| class="wikitable" | ||
| − | |+ GO ID | + | |+ GO ID Terms |
|- | |- | ||
| − | ! GO ID !! | + | ! GO ID !! Term |
|- | |- | ||
| GO:0034599 || cellular response to oxidative stress | | GO:0034599 || cellular response to oxidative stress | ||
| Line 90: | Line 76: | ||
|} | |} | ||
| − | + | ====Using YEASTRACT to Infer which Transcription Factors Regulate a Cluster of Genes==== | |
| − | |||
| − | ''' | + | # I opened the gene list in Excel for ∆CIN5 profile 22 from the stem analysis. |
| − | I | + | #* I copied the list of gene IDs. |
| + | # I launched a web browser and went to the [http://www.yeastract.com/ YEASTRACT database]. | ||
| + | #* On the left panel of the window, I clicked on the link to ''Rank by TF''. | ||
| + | #* I pasted my list of genes from my cluster into the box labeled ''ORFs/Genes''. | ||
| + | #* I checked the box for ''Check for all TFs''. | ||
| + | #* I accepted the defaults for the Regulations Filter (Documented, DNA binding or expression evidence) | ||
| + | #* I made sure to not apply a filter for "Filter Documented Regulations by environmental condition". | ||
| + | #* I ranked genes by TF using: The % of genes in the list in YEASTRACT regulated by each TF. | ||
| + | #* I clicked the ''Search'' button. | ||
| + | #* '''How many transcription factors are green or "significant"?''' | ||
| + | #**57 | ||
| + | #* I copied the table of results from the web page and pasted it into a new Excel workbook using paste values. | ||
| + | #** I uploaded [[Media:dCIN5_profile22_yeastract.xlsx|the Excel file]]. | ||
| + | #** '''Are CIN5 or GLN3 on the list? If so, what is their "% in user set", "% in YEASTRACT", and "p value"?''' | ||
| + | #*** CIN5 and GLN3 are not present on the list. | ||
| + | #** Note: Yeastract stated: <code>Unknown gene/ORF name(s), 'YCRX17W', 'YCRX18C'.</code> | ||
| + | |||
| + | ====Creating and Visualizing Your Gene Regulatory Network with GRNsight==== | ||
| + | |||
| + | # I selected 15 transcription factors from the list of "significant" transcription factors found in YEASTRACT to run the model. I added GLN3 and CIN5 if as they were not in my list. | ||
| + | #* Initially, transcription factors selection was determined by selecting the 15 most significant transcription factors by p value. The transcription factors are as follows: | ||
| + | #** Gln3 | ||
| + | #** Cin5 | ||
| + | #** Pdr3 | ||
| + | #** Rpn4 | ||
| + | #** Yap1 | ||
| + | #** Gcn4 | ||
| + | #** Rph1 | ||
| + | #** Pdr1 | ||
| + | #** Gis1 | ||
| + | #** Aft2 | ||
| + | #** Yrr1 | ||
| + | #** Mga2 | ||
| + | #** YGR067C | ||
| + | #** Sut1 | ||
| + | #** Msn4 | ||
| + | #** Stp1 | ||
| + | #** Mig1 | ||
| + | # I went to the [https://dondi.github.io/GRNsight/beta.html GRNsight beta] website. | ||
| + | # Under the "Network" panel on the left-hand side, I clicked the button "Load from database". | ||
| + | #* I typed the standard name of the transcription factor in the "Select gene" field and clicked the find button (magnifying glass). | ||
| + | #* I added transcription factors in this way until you have added the whole list above/ | ||
| + | #* I clicked the "Generate Network" button. | ||
| + | # I checked to see if all of the rectangular boxes (nodes) are connected by at least one arrow to another node. There were disconnected nodes, so I went back to the "Load from database" button and selected your transcription factors again, leaving out the nodes that were disconnected, and adding in new transcription factors down the ascending list of p values. The new list was as follows: | ||
| + | #* Gln3 | ||
| + | #* Cin5 | ||
| + | #* Pdr3 | ||
| + | #* Rpn4 | ||
| + | #* Yap1 | ||
| + | #* Gcn4 | ||
| + | #* Rph1 | ||
| + | #* Pdr1 | ||
| + | #* Aft2 | ||
| + | #* Yrr1 | ||
| + | #* YGR067C | ||
| + | #* Msn4 | ||
| + | #* Stp1 | ||
| + | #* Mig1 | ||
| + | #* Cbf1 | ||
| + | #* Bas1 | ||
| + | #* Hap2 | ||
| + | # I repeated the network generation steps again, finding that there were more disconnected nodes, until the final list of transcription factors was created (as follows): | ||
| + | #* Gln3 | ||
| + | #* Cin5 | ||
| + | #* Pdr3 | ||
| + | #* Rpn4 | ||
| + | #* Yap1 | ||
| + | #* Gcn4 | ||
| + | #* Pdr1 | ||
| + | #* Aft2 | ||
| + | #* Yrr1 | ||
| + | #* YGR067C | ||
| + | #* Stp1 | ||
| + | #* Cbf1 | ||
| + | #* Bas1 | ||
| + | #* Aft1 | ||
| + | #* Sut2 | ||
| + | #* Pho2 | ||
| + | #* Snf1 | ||
| + | #** There were 17 genes and 23 edges in my network | ||
| + | # I exported the grid layout version of the map as a PNG: | ||
| + | |||
| + | [[Image:GRN_dCIN5_prof22_hivanson_final.png|750px]] | ||
| + | Note: None of the transcriptions I tried would connect a node to CIN5. | ||
| + | |||
| + | ====Creating the GRNmap Input Workbook==== | ||
| + | |||
| + | # With your final network still open in GRNsight, I exported the data to excel. | ||
| + | #* Under "Select the Expression Data Source:", I chose "Dahlquist_2018" | ||
| + | #* Under "Select Workbook Sheets to Export:", I selected the following: | ||
| + | #** Network sheets | ||
| + | #*** "network" | ||
| + | #** Expression sheets | ||
| + | #*** dcin5_log2_expression | ||
| + | #*** dgln3_log2_expression | ||
| + | #*** wt_log2_expression | ||
| + | #** Additional sheets | ||
| + | #*** "degradation_rates" | ||
| + | #*** "optimization_parameters" | ||
| + | #*** "production_rates" | ||
| + | #*** "threshold_b" | ||
| + | #* I clicked the "Export Workbook" button. | ||
| + | # I opened the workbook in Excel to perform quality control. I checked that it has the following sheets with the following content: | ||
| + | #* The "network" sheet had an adjacency matrix with your selected regulatory transcription factors across the top row and in the first column. | ||
| + | #* The "dcin5_log2_expression", "dgln3_log2_expression", and "wt_log2_expression" sheets had log2 fold changes for each of my selected regulatory transcription factors for each time point (15, 30, 60, 90, 120). | ||
| + | #* The "production_rates" and "degradation_rates" sheets had values for each gene. | ||
| + | #* The "threshold_b" sheet had a value of 0 for each gene. | ||
| + | #* In the "optimization_parameters" sheet, I changed the "alpha" value to 0.02 instead of 0.002. | ||
| + | #* I inserted a new worksheet and named it "network_weights". | ||
| + | #** I copied the entire content of the "network" sheet into the "network_weights" sheet. | ||
| + | # I saved and uploaded [[Media:GRN_dCIN5_profile22_map_hivanson.xlsx|my Excel Workbook]] | ||
===Data & Files=== | ===Data & Files=== | ||
| Line 105: | Line 200: | ||
[[Media:DCIN5_profiles_GOlist_hivanson.zip|Stem dCIN5 significant profiles GOlist folder]] | [[Media:DCIN5_profiles_GOlist_hivanson.zip|Stem dCIN5 significant profiles GOlist folder]] | ||
| + | |||
| + | [[Media:dCIN5_profile22_yeastract.xlsx|dCIN5 Profile 22 genelist in yeastract results]] | ||
[[Media:BIOL367_S24_hivanson_dCIN5_p-value_slide.pptx|Slides]] | [[Media:BIOL367_S24_hivanson_dCIN5_p-value_slide.pptx|Slides]] | ||
| + | |||
| + | [[Media:GRN_dCIN5_profile22_map_hivanson.xlsx|GRNsight data for dCIN5 profile 22]] | ||
===Conclusion=== | ===Conclusion=== | ||
| + | The analysis of the ∆CIN5 DNA microarray set revealed that profile 22 had significantly more genes than predicted that followed the trend of spiking in upregulation at the 90-minute time mark of cold shock. The genes found in this profile are associated with cell stress responses, as well as various cell parts and organelles. A gene regulatory network was formed of the most significantly affected (by cold shock) transcription factors in profile 22, and CIN5 did not connect into the network with the most significant transcription factors. Further analysis will be performed after my workbook is run through GRNmap modeling software. | ||
| + | |||
===Acknowledgments=== | ===Acknowledgments=== | ||
| + | I worked on this assignment in class on 3/28/2024 and 4/2/2024 under the guidance of [[User:Kdahlquist|Dr. Kam Dahlquist]]. All procedure was edited from [https://xmlpipedb.cs.lmu.edu/biodb/spring2024/index.php/Week_10 LMU Bio DB Week 10]. | ||
| + | |||
| + | Except for what is noted above, this individual journal entry was completed by me and not copied from another source. | ||
| + | |||
| + | [[User:Hivanson|Hivanson]] ([[User talk:Hivanson|talk]]) 21:13, 3 April 2024 (PDT) | ||
| + | |||
===References=== | ===References=== | ||
| + | *Gene Ontology Resource. (n.d.). Gene Ontology Resource. Retrieved April 3, 2024, from http://geneontology.org/ | ||
| + | *GRNsight—Beta. (n.d.). Retrieved April 3, 2024, from https://dondi.github.io/GRNsight/beta.html | ||
*Help:Table. (2024). In Wikipedia. Retrieved March 25, 2024, from https://en.wikipedia.org/w/index.php?title=Help:Table&oldid=1213890372 | *Help:Table. (2024). In Wikipedia. Retrieved March 25, 2024, from https://en.wikipedia.org/w/index.php?title=Help:Table&oldid=1213890372 | ||
| + | *LMU BioDB 2024. (2024). Week 10. Retrieved April 3, 2024, from https://xmlpipedb.cs.lmu.edu/biodb/spring2024/index.php/Week_10 | ||
| + | *Saccharomyces Genome Database. (n.d.) CIN5 / YOR028C Overview. Retrieved April 3, 2024, from https://www.yeastgenome.org/locus/S000005554 | ||
| + | *S.cerevisiae—Yeastract. (n.d.). Retrieved April 3, 2024, from http://www.yeastract.com/ | ||
| + | *STEM: Short Time-series Expression Miner. (n.d.). Retrieved April 3, 2024, from https://www.cs.cmu.edu/~jernst/stem/ | ||
| + | |||
{{Template:Hivanson}} | {{Template:Hivanson}} | ||
Latest revision as of 22:11, 3 April 2024
Purpose
We are analyzing a DNA microarray dataset, involving clustering and analysis of gene regulatory networks using GRNsight. The DNA microarray dataset is of the S. cerevisiae genome's cold shock response in a ∆CIN5 strain. This analysis will enable us to understand the role of the transcription factor CIN5's role in the cold shock response in S. cerevisiae.
Methods/Results
Clustering and GO Term Enrichment with stem
- Preparing microarray data file for loading into STEM.
- I inserted a new worksheet into my Excel workbook, and named it "dCIN5_stem".
- I selected all of the data from the "ANOVA_dCIN5" worksheet and special pasted values into the "dCIN5_stem" worksheet.
- I renamed Column A to "SPOT" and Column B to "Gene Symbol." I deleted the column named "Standard_Name."
- I filtered the data in "dCIN5_B-H_p-value" > 0.05.
- Once the data has been filtered, I selected and deleted all of the rows (except for your header row). I undid the filter.
- I deleted all of the data columns except for the Average Log Fold change columns for timepoints 15m, 30m, 60m, 90m, and 120m.
- I renamed the data columns from "dCIN5_AvgLogFC_t
- I removed #DIV/0! errors using find and replace with nothing in the replace field.
- I saved the dCIN5_stem sheet as Text (Tab-delimited) (*.txt).
- I downloaded and extracted the STEM software. Click here to go to the STEM web site.
- I downloaded the Gene Ontology and yeast GO annotations from the 10 Wiki page.
- I launched the STEM program.
- Running STEM
- In section 1 (Expression Data Info) of the the main STEM interface window, I clicked on the Browse... button to navigate to and select the .txt file.
- I clicked on the radio button No normalization/add 0.
- I checked the box next to Spot IDs included in the data file.
- In section 2 (Gene Info) of the main STEM interface window, I left the default selection for the three drop-down menu selections for Gene Annotation Source, Cross Reference Source, and Gene Location Source as "User provided".
- I clicked the "Browse..." button to the right of the "Gene Annotation File" item. I selected the file "gene_association.sgd.gz" and click Open.
- In section 3 (Options) of the main STEM interface window, I ensured that the Clustering Method says "STEM Clustering Method" and did not change the defaults for Maximum Number of Model Profiles or Maximum Unit Change in Model Profiles between Time Points.
- In section 4 (Execute) I clicked on the yellow Execute button to run STEM.
- In section 1 (Expression Data Info) of the the main STEM interface window, I clicked on the Browse... button to navigate to and select the .txt file.
- Viewing and Saving STEM Results
- A new window opened called "All STEM Profiles (1)".
- I clicked on the button that says "Interface Options...". At the bottom of the Interface Options window that appears below where it says "X-axis scale should be:", I clicked on the radio button that says "Based on real time". I then closed the Interface Options window.
- I screenshotted this window and pasted it into my PowerPoint presentation.
- I clicked on each of the significant/colored profiles to open a window showing a more detailed plot containing all of the genes in that profile.
- I screenshoted each of the individual profile windows and saved the images in my PowerPoint presentation.
- For each of the profiles, I clicked on the "Profile Gene Table" button, then clicked on the "Save Table" button to save the file for each profile. This can be found in the Data and Files section below.
- For each of the significant profiles, I clicked on the "Profile GO Table" to see the list of Gene Ontology terms belonging to the profile, then clicked on the "Save Table" button for each profile. This can be found in the Data and Files section below.
- A new window opened called "All STEM Profiles (1)".
- Analyzing and Interpreting STEM Results
- I selected profile 22 for further intepretation of the data.
- Why did you select this profile? In other words, why was it interesting to you?
- I selected profile 22 because the overall trend of a spike only at 90 minutes is different from the rest of the profiles' trends. Further, it had the second highest number of genes belonging to it out of all significant profiles.
- How many genes belong to this profile?
- 179 genes
- How many genes were expected to belong to this profile?
- 22.9 genes
- What is the p value for the enrichment of genes in this profile?
- 3.4E-98
- I opened the GO list file that I saved for profile 22 in Excel. I selected the third row and then filtered on the "p-value" column to show only GO terms that have a p value of < 0.05.
- How many GO terms are associated with this profile at p < 0.05?
- 25
- I filtered on the "Corrected p-value" column to show only GO terms that have a corrected p value of < 0.05.
- How many GO terms are associated with this profile with a corrected p value < 0.05?
- 7
- I select 6 Gene Ontology terms from your filtered list
- These were selected by choosing the six most significant terms that were not redundant with each other.
- I looked up the definitions for each of the terms at http://geneontology.org.
- Why does the cell react to cold shock by changing the expression of genes associated with these GO terms?
- The genes that are associated with these GO terms include stress responses, like
cellular response to oxidative stress, and its related terms that I excluded from the above table due to similarity. This makes sense as cold shock is considered a stressor. Additionally, various cellular structure components are associated with these GO terms, such ascytoplasm,cytoskeleton, andfungal-type vacuolewhich all may need to have regulated rigidity, fluidity or some other physical characteristics at lower temperatures.
- The genes that are associated with these GO terms include stress responses, like
- Also, what does this have to do with the transcription factor being deleted (for the groups working with deletion strain data)
- I want to compare our results to that of the wildtype. The SGD description of CIN5 says it is
nuclearly localized under oxidative stress, so perhaps with the deletion of CIN5 transcription factor, oxidative stress, and possibly other stress responses would be unable to occur.
- I want to compare our results to that of the wildtype. The SGD description of CIN5 says it is
- Why did you select this profile? In other words, why was it interesting to you?
- I selected profile 22 for further intepretation of the data.
| GO ID | Term |
|---|---|
| GO:0034599 | cellular response to oxidative stress |
| GO:0005737 | cytoplasm |
| GO:0006897 | endocytosis |
| GO:0030479 | actin cortical patch |
| GO:0005856 | cytoskeleton |
| GO:0000324 | fungal-type vacuole |
Using YEASTRACT to Infer which Transcription Factors Regulate a Cluster of Genes
- I opened the gene list in Excel for ∆CIN5 profile 22 from the stem analysis.
- I copied the list of gene IDs.
- I launched a web browser and went to the YEASTRACT database.
- On the left panel of the window, I clicked on the link to Rank by TF.
- I pasted my list of genes from my cluster into the box labeled ORFs/Genes.
- I checked the box for Check for all TFs.
- I accepted the defaults for the Regulations Filter (Documented, DNA binding or expression evidence)
- I made sure to not apply a filter for "Filter Documented Regulations by environmental condition".
- I ranked genes by TF using: The % of genes in the list in YEASTRACT regulated by each TF.
- I clicked the Search button.
- How many transcription factors are green or "significant"?
- 57
- I copied the table of results from the web page and pasted it into a new Excel workbook using paste values.
- I uploaded the Excel file.
- Are CIN5 or GLN3 on the list? If so, what is their "% in user set", "% in YEASTRACT", and "p value"?
- CIN5 and GLN3 are not present on the list.
- Note: Yeastract stated:
Unknown gene/ORF name(s), 'YCRX17W', 'YCRX18C'.
Creating and Visualizing Your Gene Regulatory Network with GRNsight
- I selected 15 transcription factors from the list of "significant" transcription factors found in YEASTRACT to run the model. I added GLN3 and CIN5 if as they were not in my list.
- Initially, transcription factors selection was determined by selecting the 15 most significant transcription factors by p value. The transcription factors are as follows:
- Gln3
- Cin5
- Pdr3
- Rpn4
- Yap1
- Gcn4
- Rph1
- Pdr1
- Gis1
- Aft2
- Yrr1
- Mga2
- YGR067C
- Sut1
- Msn4
- Stp1
- Mig1
- Initially, transcription factors selection was determined by selecting the 15 most significant transcription factors by p value. The transcription factors are as follows:
- I went to the GRNsight beta website.
- Under the "Network" panel on the left-hand side, I clicked the button "Load from database".
- I typed the standard name of the transcription factor in the "Select gene" field and clicked the find button (magnifying glass).
- I added transcription factors in this way until you have added the whole list above/
- I clicked the "Generate Network" button.
- I checked to see if all of the rectangular boxes (nodes) are connected by at least one arrow to another node. There were disconnected nodes, so I went back to the "Load from database" button and selected your transcription factors again, leaving out the nodes that were disconnected, and adding in new transcription factors down the ascending list of p values. The new list was as follows:
- Gln3
- Cin5
- Pdr3
- Rpn4
- Yap1
- Gcn4
- Rph1
- Pdr1
- Aft2
- Yrr1
- YGR067C
- Msn4
- Stp1
- Mig1
- Cbf1
- Bas1
- Hap2
- I repeated the network generation steps again, finding that there were more disconnected nodes, until the final list of transcription factors was created (as follows):
- Gln3
- Cin5
- Pdr3
- Rpn4
- Yap1
- Gcn4
- Pdr1
- Aft2
- Yrr1
- YGR067C
- Stp1
- Cbf1
- Bas1
- Aft1
- Sut2
- Pho2
- Snf1
- There were 17 genes and 23 edges in my network
- I exported the grid layout version of the map as a PNG:
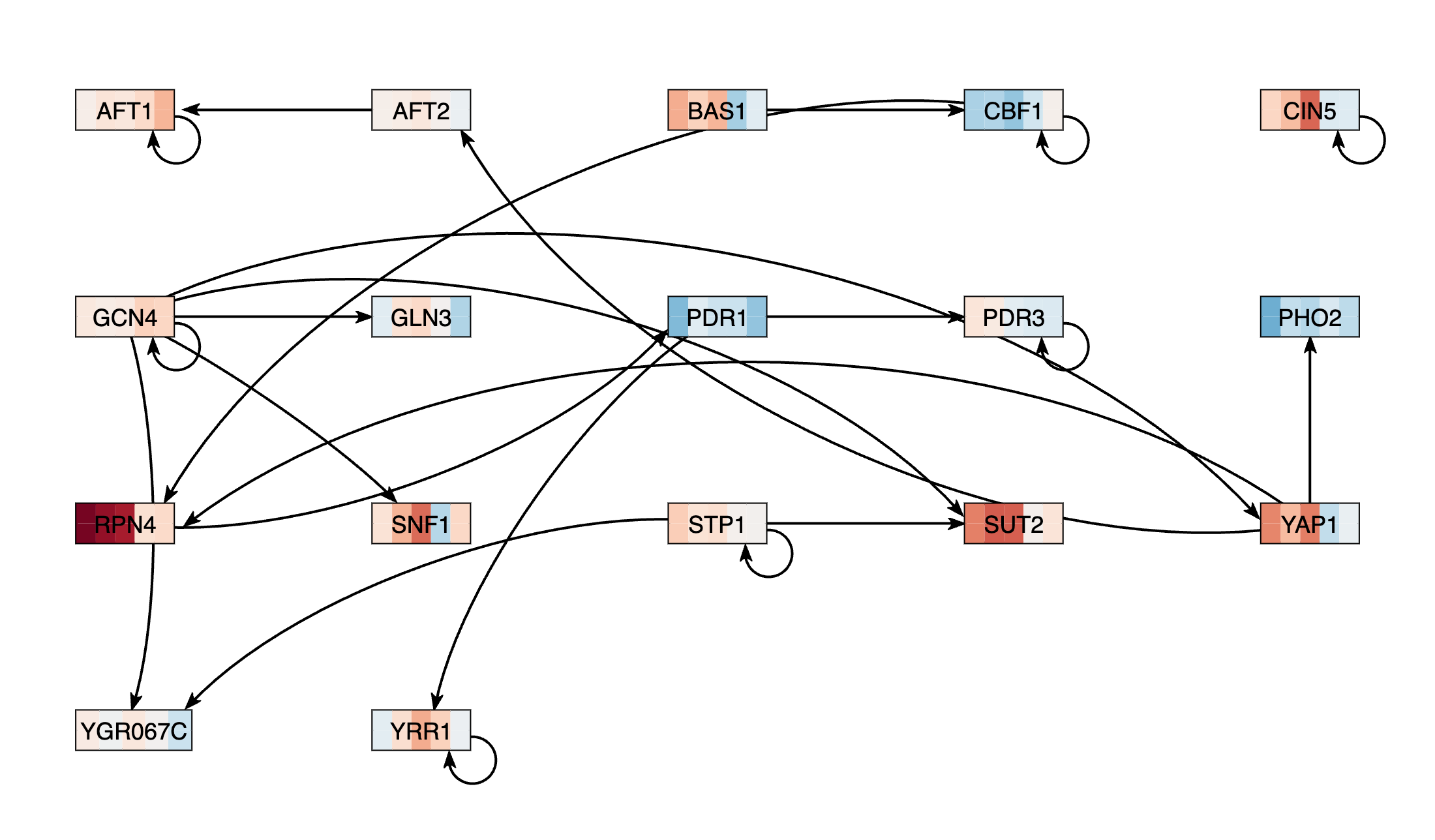 Note: None of the transcriptions I tried would connect a node to CIN5.
Note: None of the transcriptions I tried would connect a node to CIN5.
Creating the GRNmap Input Workbook
- With your final network still open in GRNsight, I exported the data to excel.
- Under "Select the Expression Data Source:", I chose "Dahlquist_2018"
- Under "Select Workbook Sheets to Export:", I selected the following:
- Network sheets
- "network"
- Expression sheets
- dcin5_log2_expression
- dgln3_log2_expression
- wt_log2_expression
- Additional sheets
- "degradation_rates"
- "optimization_parameters"
- "production_rates"
- "threshold_b"
- Network sheets
- I clicked the "Export Workbook" button.
- I opened the workbook in Excel to perform quality control. I checked that it has the following sheets with the following content:
- The "network" sheet had an adjacency matrix with your selected regulatory transcription factors across the top row and in the first column.
- The "dcin5_log2_expression", "dgln3_log2_expression", and "wt_log2_expression" sheets had log2 fold changes for each of my selected regulatory transcription factors for each time point (15, 30, 60, 90, 120).
- The "production_rates" and "degradation_rates" sheets had values for each gene.
- The "threshold_b" sheet had a value of 0 for each gene.
- In the "optimization_parameters" sheet, I changed the "alpha" value to 0.02 instead of 0.002.
- I inserted a new worksheet and named it "network_weights".
- I copied the entire content of the "network" sheet into the "network_weights" sheet.
- I saved and uploaded my Excel Workbook
Data & Files
Excel file with microarray data for dCIN5
Tab text file with dCIN5 stem data
Stem dCIN5 significant profiles genelist folder
Stem dCIN5 significant profiles GOlist folder
dCIN5 Profile 22 genelist in yeastract results
GRNsight data for dCIN5 profile 22
Conclusion
The analysis of the ∆CIN5 DNA microarray set revealed that profile 22 had significantly more genes than predicted that followed the trend of spiking in upregulation at the 90-minute time mark of cold shock. The genes found in this profile are associated with cell stress responses, as well as various cell parts and organelles. A gene regulatory network was formed of the most significantly affected (by cold shock) transcription factors in profile 22, and CIN5 did not connect into the network with the most significant transcription factors. Further analysis will be performed after my workbook is run through GRNmap modeling software.
Acknowledgments
I worked on this assignment in class on 3/28/2024 and 4/2/2024 under the guidance of Dr. Kam Dahlquist. All procedure was edited from LMU Bio DB Week 10.
Except for what is noted above, this individual journal entry was completed by me and not copied from another source.
Hivanson (talk) 21:13, 3 April 2024 (PDT)
References
- Gene Ontology Resource. (n.d.). Gene Ontology Resource. Retrieved April 3, 2024, from http://geneontology.org/
- GRNsight—Beta. (n.d.). Retrieved April 3, 2024, from https://dondi.github.io/GRNsight/beta.html
- Help:Table. (2024). In Wikipedia. Retrieved March 25, 2024, from https://en.wikipedia.org/w/index.php?title=Help:Table&oldid=1213890372
- LMU BioDB 2024. (2024). Week 10. Retrieved April 3, 2024, from https://xmlpipedb.cs.lmu.edu/biodb/spring2024/index.php/Week_10
- Saccharomyces Genome Database. (n.d.) CIN5 / YOR028C Overview. Retrieved April 3, 2024, from https://www.yeastgenome.org/locus/S000005554
- S.cerevisiae—Yeastract. (n.d.). Retrieved April 3, 2024, from http://www.yeastract.com/
- STEM: Short Time-series Expression Miner. (n.d.). Retrieved April 3, 2024, from https://www.cs.cmu.edu/~jernst/stem/
- Hivanson
- Hivanson Week 1 | Week 1 Assignment
- Hivanson Week 2 | Week 2 Assignment
- IMD3 Hivanson and Nstojan1 Week 3 | Week 3 Assignment
- NeMO Week 4 | Week 4 Assignment
- Hivanson Week 5 | Week 5 Assignment
- Hivanson Week 6 | Week 6 Assignment
- Hivanson Week 8 | Week 8 Assignment
- Hivanson Week 9 | Week 9 Assignment
- Hivanson Week 10 | Week 10 Assignment
- Hivanson Week 12 | Week 12 Assignment
- Hivanson Week 13 | Week 13 Assignment
- Hivanson Week 14 | Week 14 Assignment
- Hivanson Week 15 | Week 15 Assignment
- Main page