Difference between revisions of "Asandle1 Week 5"
(→Assignment Questions: added questions without answers) |
(→Andrew Sandler's References: fixed numbering) |
||
| (17 intermediate revisions by the same user not shown) | |||
| Line 13: | Line 13: | ||
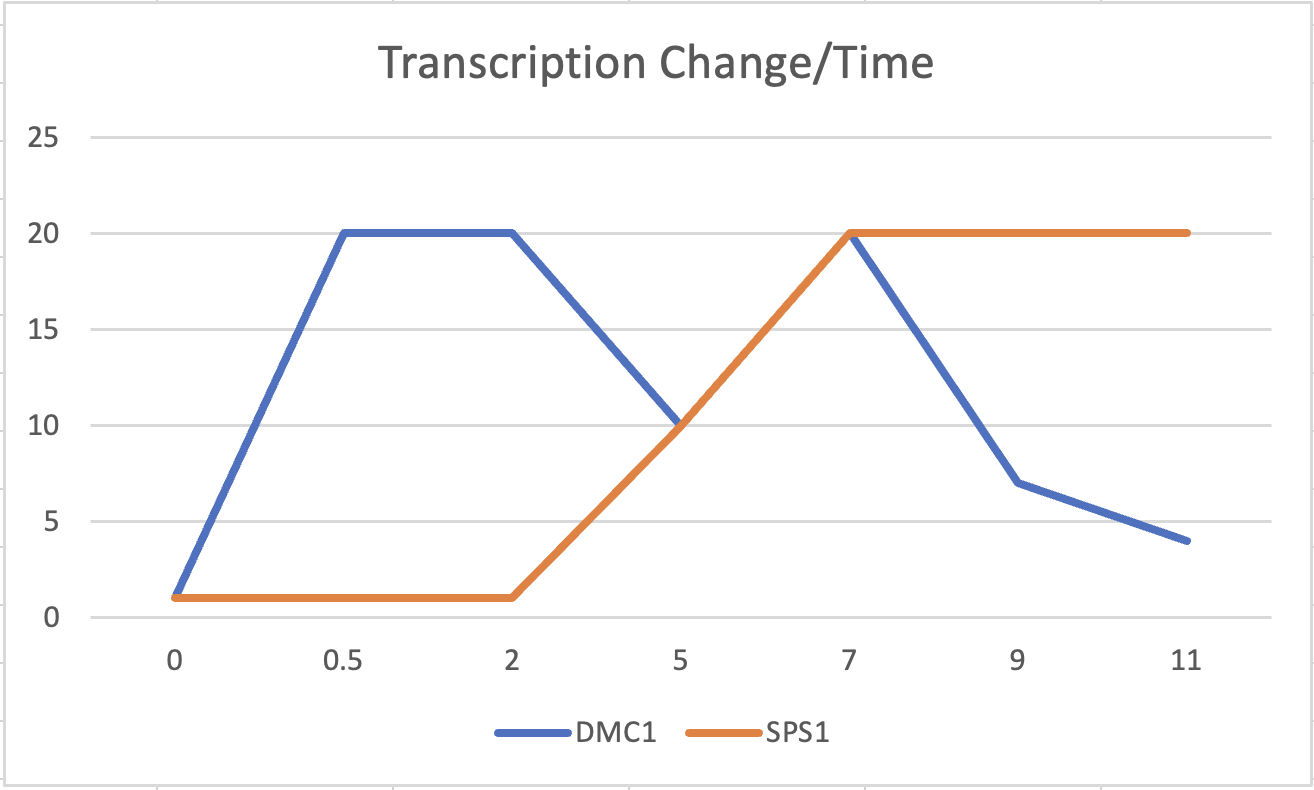
Choose two genes from Figure 4.6b (PDF of figures on Brightspace) and draw a graph to represent the change in transcription over time. You can either create your plot in Excel and put the image up on your wiki page or you can do it by hand and upload a picture or scan. | Choose two genes from Figure 4.6b (PDF of figures on Brightspace) and draw a graph to represent the change in transcription over time. You can either create your plot in Excel and put the image up on your wiki page or you can do it by hand and upload a picture or scan. | ||
| + | |||
| + | [[File:Asandle1_Transcription_Change_Over_Time_Week5.png|600px|This Graph answer Question 5]] | ||
<b>(Question 6b, p. 110)</b> | <b>(Question 6b, p. 110)</b> | ||
| + | Look at Figure 4.7, which depicts the loss of oxygen over time and the transcriptional response of three genes. These data are the ratios of transcription for genes X, Y, and Z during the depletion of oxygen. Using the color scale from Figure 4.6, determine the color for each ratio in Figure 4.7b. (Use the nomenclature "bright green", "medium green", "dim green", "black", "dim red", "medium red", or "bright red" for your answers.) | ||
| + | X: Black, Dim Red, Black, Medium Green | ||
| + | Y: Black, Medium Red, Dim Green, Bright Green | ||
| + | Z: Black, Dim Red, Dim Red, Dim Red | ||
<b>(Question 7, p. 110)</b> | <b>(Question 7, p. 110)</b> | ||
| + | |||
| + | Were any of the genes in Figure 4.7b transcribed similarly? If so, which ones were transcribed similarly to which ones? | ||
| + | |||
| + | Yes, X and Y, my explanation is this below: | ||
| + | The genes all have ratios of 1.0 an hour in so they are not showing induction or repression yet. Then an hour in they have all been inducting but at different rates. Then 5 hours in they all split in what they are doing, Gene X is having no change in transcription, while Gene Y is starting to repress very slightly and gene Z is inducting again. Then at 9 hours, Gene X and Y are repressing a lot. All in all Gene X and Y are the most similar in terms of their transcription ratios here. | ||
| + | |||
<b>(Question 9, p. 118)</b> | <b>(Question 9, p. 118)</b> | ||
| + | |||
| + | Why would most spots be yellow at the first time point? I.e., what is the technical reason that spots show up as yellow - where does the yellow color come from? And, what would be the biological reason that the experiment resulted in most spots being yellow? | ||
| + | |||
| + | Most would be yellow in the first time point because there is no difference in levels of expression at the start. They show up yellow because one gene is using the green fluorescence and one is using the red, so they look yellow on the computer screen when they are imposed on one another. If two genes were expressed equally that would be the reason for an experiment to result in most spots being yellow. | ||
| + | |||
<b>(Question 10, p. 118)</b> | <b>(Question 10, p. 118)</b> | ||
| + | |||
| + | Go to the [http://www.yeastgenome.org ''Saccharomyces'' Genome Database] and search for the gene TEF4; you will see it is involved in translation. Look at the time point labeled OD 3.7 in Figure 4.12, and find the TEF4 spot. Over the course of this experiment, was TEF4 induced or repressed? Hypothesize why TEF4’s change in expression was part of the cell’s response to a reduction in available glucose (i.e., the only available food). | ||
| + | |||
| + | At the beginning the TEF4 is yellow, this shows it is neither being induced or repressed compared to the control. As the time continues and the available glucose is reduced, we see it slowly becoming greener which means it is being more repressed than the control. According to [https://www.frontiersin.org/articles/10.3389/fmolb.2021.816398/full this journal article], Translation Elongation Factors are a key part of protein synthesis. So it seems possible that with less glucose available the cells are doing less and therefore need to create less YKL081W Protein. Which is what it says the protein it creates is called on the Saccharomyces Genome Database. | ||
| + | |||
<b>(Question 11, p. 120)</b> | <b>(Question 11, p. 120)</b> | ||
| + | |||
| + | Why would TCA cycle genes be induced if the glucose supply is running out? | ||
| + | |||
| + | I used [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC151572/#:~:text=Its%20main%20catalytic%20function%20is,acids%2C%20heme%2C%20and%20glucose. this online paper] as my source. It looks like they might be induced to help use energy more efficiently or to use alternate energy sources. | ||
| + | |||
<b>(Question 12, p. 120)</b> | <b>(Question 12, p. 120)</b> | ||
| + | |||
| + | What mechanism could the genome use to ensure genes for enzymes in a common pathway are induced or repressed simultaneously? | ||
| + | |||
| + | The genome could use specific transcription factors as that is what we already see in the reading in figure 4.14. | ||
| + | |||
<b>(Question 13, p. 121)</b> | <b>(Question 13, p. 121)</b> | ||
| + | |||
| + | Consider a microarray experiment where cells deleted for the repressor TUP1 were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented glucose-repressed genes to be in the later time points of this experiment? | ||
| + | |||
| + | The genes for glucose would be highly induced and therefore red. | ||
| + | My reasoning is below in image form, I made a table to think through this on my notes. | ||
| + | |||
| + | [[File:Table_Glucose_TUP1_Repression.png|600px|thumb| This is the table that shows my reasoning for answering the question regarding TUP1]] | ||
| + | |||
| + | Regarding question 13: [[user:Kdahlquist| Dr. Dahlquist]] | ||
| + | How do we know that the specific genes aren't already highly induced and they are only turned off because of the strong repression, and therefor by removing the repression at the start they are already immediately strongly induced? | ||
<b>(Question 14, p. 121)</b> | <b>(Question 14, p. 121)</b> | ||
| + | |||
| + | Consider a microarray experiment where cells that overexpress the transcription factor Yap1p were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented Yap1p target genes to be in the later time points of this experiment? | ||
| + | |||
| + | They would be green since Yap1p represses its target genes and therefore over expressing would cause more repression of the downstream genes. | ||
| + | |||
<b>(Question 16, p. 121)</b> | <b>(Question 16, p. 121)</b> | ||
| + | |||
| + | Using the microarray data, how could you verify that you had truly deleted TUP1 or overexpressed YAP1 in the experiments described in questions 8 and 9? | ||
| + | |||
| + | You would just look at the genes that are known to be repressed by TUP1 under normal conditions and confirm they are being highly induced.You would do the opposite for YAP1 and confirm that the Genes downstream are being repressed. | ||
| + | |||
| + | ===Andrew Sandler's References=== | ||
| + | |||
| + | <b>Bibliography</b> | ||
| + | |||
| + | #LMU BioDB 2024. (2024). Week 5. Retrieved February 15, 2024, from https://xmlpipedb.cs.lmu.edu/biodb/Spring2024/index.php/Week_5 | ||
| + | #McCammon, M. T., Epstein, C. B., Przybyla-Zawislak, B., McAlister-Henn, L., & Butow, R. A. (2003, March). Global transcription analysis of Krebs tricarboxylic acid cycle mutants reveals an alternating pattern of gene expression and effects on hypoxic and oxidative genes. Molecular biology of the cell. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC151572/#:~:text=Its%20main%20catalytic%20function%20is,acids%2C%20heme%2C%20and%20glucose. | ||
| + | #TEF4 / YKL081W Protein. TEF4 Protein | SGD. (n.d.-a). https://www.yeastgenome.org/locus/S000001564/protein | ||
| + | #Xu, B., Liu, L., & Song, G. (2021, December 20). Functions and regulation of translation elongation factors. Frontiers. https://www.frontiersin.org/articles/10.3389/fmolb.2021.816398/full | ||
| + | #Campbell, A.M. and Heyer, L.J. (2003), “Chapter 4: Basic Research with DNA Microarrays”, in Discovering Genomics, Proteomics, and Bioinformatics, Cold Spring Harbor Laboratory Press, pp. 107-124. | ||
| + | |||
| + | |||
| + | <b>Acknowledgements</b> | ||
| + | |||
| + | - I worked on this assignment on my own. | ||
| + | |||
| + | - I used only my own wiki and html knowledge | ||
| + | |||
| + | - I copied my citation for Campbell article from Katie's page. | ||
| + | |||
| + | - All work is my own except where acknowledged otherwise [[User:Asandle1|Asandle1]] ([[User talk:Asandle1|talk]]) 19:36, 15 February 2024 (PST) | ||
| + | |||
| + | -Except for what is noted above, this individual journal entry was completed by me and not copied from another source. [[User:Asandle1|Asandle1]] ([[User talk:Asandle1|talk]]) 19:46, 15 February 2024 (PST) | ||
==Delete This Sentence before submitting reminder Category is last thing on page== | ==Delete This Sentence before submitting reminder Category is last thing on page== | ||
[[Category:Journal Entry]] | [[Category:Journal Entry]] | ||
Latest revision as of 19:49, 15 February 2024
To User Page: User: Asandle1 To Template: Template:Asandle1
Contents
Assignment Pages
Journals
Individual
Class Journals
Electronic Lab Notebook
Purpose
The purpose of this assignment is to learn about DNA microarrays. It also is a way to learn about data and how we can organize data to derive meaningful information from experimental results.
Assignment Questions
(Question 5, p. 110)
Choose two genes from Figure 4.6b (PDF of figures on Brightspace) and draw a graph to represent the change in transcription over time. You can either create your plot in Excel and put the image up on your wiki page or you can do it by hand and upload a picture or scan.
(Question 6b, p. 110)
Look at Figure 4.7, which depicts the loss of oxygen over time and the transcriptional response of three genes. These data are the ratios of transcription for genes X, Y, and Z during the depletion of oxygen. Using the color scale from Figure 4.6, determine the color for each ratio in Figure 4.7b. (Use the nomenclature "bright green", "medium green", "dim green", "black", "dim red", "medium red", or "bright red" for your answers.)
X: Black, Dim Red, Black, Medium Green Y: Black, Medium Red, Dim Green, Bright Green Z: Black, Dim Red, Dim Red, Dim Red
(Question 7, p. 110)
Were any of the genes in Figure 4.7b transcribed similarly? If so, which ones were transcribed similarly to which ones?
Yes, X and Y, my explanation is this below: The genes all have ratios of 1.0 an hour in so they are not showing induction or repression yet. Then an hour in they have all been inducting but at different rates. Then 5 hours in they all split in what they are doing, Gene X is having no change in transcription, while Gene Y is starting to repress very slightly and gene Z is inducting again. Then at 9 hours, Gene X and Y are repressing a lot. All in all Gene X and Y are the most similar in terms of their transcription ratios here.
(Question 9, p. 118)
Why would most spots be yellow at the first time point? I.e., what is the technical reason that spots show up as yellow - where does the yellow color come from? And, what would be the biological reason that the experiment resulted in most spots being yellow?
Most would be yellow in the first time point because there is no difference in levels of expression at the start. They show up yellow because one gene is using the green fluorescence and one is using the red, so they look yellow on the computer screen when they are imposed on one another. If two genes were expressed equally that would be the reason for an experiment to result in most spots being yellow.
(Question 10, p. 118)
Go to the Saccharomyces Genome Database and search for the gene TEF4; you will see it is involved in translation. Look at the time point labeled OD 3.7 in Figure 4.12, and find the TEF4 spot. Over the course of this experiment, was TEF4 induced or repressed? Hypothesize why TEF4’s change in expression was part of the cell’s response to a reduction in available glucose (i.e., the only available food).
At the beginning the TEF4 is yellow, this shows it is neither being induced or repressed compared to the control. As the time continues and the available glucose is reduced, we see it slowly becoming greener which means it is being more repressed than the control. According to this journal article, Translation Elongation Factors are a key part of protein synthesis. So it seems possible that with less glucose available the cells are doing less and therefore need to create less YKL081W Protein. Which is what it says the protein it creates is called on the Saccharomyces Genome Database.
(Question 11, p. 120)
Why would TCA cycle genes be induced if the glucose supply is running out?
I used this online paper as my source. It looks like they might be induced to help use energy more efficiently or to use alternate energy sources.
(Question 12, p. 120)
What mechanism could the genome use to ensure genes for enzymes in a common pathway are induced or repressed simultaneously?
The genome could use specific transcription factors as that is what we already see in the reading in figure 4.14.
(Question 13, p. 121)
Consider a microarray experiment where cells deleted for the repressor TUP1 were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented glucose-repressed genes to be in the later time points of this experiment?
The genes for glucose would be highly induced and therefore red. My reasoning is below in image form, I made a table to think through this on my notes.
Regarding question 13: Dr. Dahlquist How do we know that the specific genes aren't already highly induced and they are only turned off because of the strong repression, and therefor by removing the repression at the start they are already immediately strongly induced?
(Question 14, p. 121)
Consider a microarray experiment where cells that overexpress the transcription factor Yap1p were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented Yap1p target genes to be in the later time points of this experiment?
They would be green since Yap1p represses its target genes and therefore over expressing would cause more repression of the downstream genes.
(Question 16, p. 121)
Using the microarray data, how could you verify that you had truly deleted TUP1 or overexpressed YAP1 in the experiments described in questions 8 and 9?
You would just look at the genes that are known to be repressed by TUP1 under normal conditions and confirm they are being highly induced.You would do the opposite for YAP1 and confirm that the Genes downstream are being repressed.
Andrew Sandler's References
Bibliography
- LMU BioDB 2024. (2024). Week 5. Retrieved February 15, 2024, from https://xmlpipedb.cs.lmu.edu/biodb/Spring2024/index.php/Week_5
- McCammon, M. T., Epstein, C. B., Przybyla-Zawislak, B., McAlister-Henn, L., & Butow, R. A. (2003, March). Global transcription analysis of Krebs tricarboxylic acid cycle mutants reveals an alternating pattern of gene expression and effects on hypoxic and oxidative genes. Molecular biology of the cell. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC151572/#:~:text=Its%20main%20catalytic%20function%20is,acids%2C%20heme%2C%20and%20glucose.
- TEF4 / YKL081W Protein. TEF4 Protein | SGD. (n.d.-a). https://www.yeastgenome.org/locus/S000001564/protein
- Xu, B., Liu, L., & Song, G. (2021, December 20). Functions and regulation of translation elongation factors. Frontiers. https://www.frontiersin.org/articles/10.3389/fmolb.2021.816398/full
- Campbell, A.M. and Heyer, L.J. (2003), “Chapter 4: Basic Research with DNA Microarrays”, in Discovering Genomics, Proteomics, and Bioinformatics, Cold Spring Harbor Laboratory Press, pp. 107-124.
Acknowledgements
- I worked on this assignment on my own.
- I used only my own wiki and html knowledge
- I copied my citation for Campbell article from Katie's page.
- All work is my own except where acknowledged otherwise Asandle1 (talk) 19:36, 15 February 2024 (PST)
-Except for what is noted above, this individual journal entry was completed by me and not copied from another source. Asandle1 (talk) 19:46, 15 February 2024 (PST)