Asandle1 Week 10
Contents
Methods/Results/Electronic Notebook
Purpose
The purpose of this assignment is to get experience with the analysis part of the "data life cycle" for DNA microarray datasets and to "develop an intuition about different p-value cut-offs and their meanings.
It also is to show how to keep a detailed electronic lab notebook to make our research reproducible to develop an intuition about what different p-value cut-offs mean.
Methods/Electronic Notebook
Part 1: Data Setup and STEM Installation
1) I inserted a new worksheet into my Excel workbook and named it dGLN3_stem.
2) Selected all of the data from my dGLN3_ANOVA worksheet and pasted it as values into my new dGLN3_stem worksheet.
3) Renamed my left MasterIndex column to SPOT
4) Renamed Column B to "Gene Symbol".
5) Deleted the column named "Standard_Name".
6) Filtered the data on the B-H corrected p value to be > 0.05.
7) Selected all of the rows (except for the header) and deleted the rows by right-clicking and choosing "Delete Row" from the context menu.
8)Undid the filter.
9) Deleted all of the data columns EXCEPT for the Average Log Fold change columns for each timepoint (for example, wt_AvgLogFC_t15, etc.).
10) Helped Charlotte Kaplan do the same filtering step. She had a small issue where she deleted the data on the rows but not the rows.
11) Renamed the data columns with just the time and units (for example, 15m, 30m, etc.).
12) Helped Charlotte with Pasting Values.
13) There were some #DIV/0! errors. To fix, I am opening the Find/Replace dialog and searching for the DIV errors by #DIV/0!.
14) It removed 110 DIV/0’s.
15) Saved my work. Then used Save As to save it as .txt file.
16) I did not turn on file extensions because I am on a mac and it shows me already.
17) Tried to download the Stem software on mac but it was not a quick process so I moved to the Windows computer.
18) I used Box to upload the spreadsheet that I saved as a text file.
19) I downloaded the text file onto the windows computer.
20) I extracted it.
21) Dr. Dahlquist helped me turn on file extensions.
22) I downloaded and extracted the STEM software onto the computer.
23) I downloaded the Gene Ontology file gene_ontology.obo from https://lmu.box.com/s/t8i5s1z1munrcfxzzs7nv7q2edsktxgl
24) I downloaded the gene_association.sgd.gz file from https://lmu.box.com/s/zlr1s8fjogfssa1wl59d5shyybtm1d49
25) I put the files into the STEM folder that was created when I extracted all from the Stem.zip earlier.
Part 2: Using STEM
1) I double clicked on stem.jar to run the program
2) STEM opened into the main interface window and I clicked on the Browse button to navigate to my file AS&CK_BIOL367_STEM.txt and selected it.
3) I clicked on the button that says No normalization/add 0.
4) I clicked on the box next to Spot IDs included in the data file.
5) I did not make any changes to the default selection for the three drop-down menu selections for Gene Annotation Source, Cross Reference Source, and Gene Location Source. They are all still set as "User Provided".
6) I clicked the Browse button on the right of the "Gene Annotation File" item.
7) I looked in the "stem" folder and selected the file called "gene_association.sdg.gz" which I had previously downloaded and put into the "stem" folder.
8) In the third section of the STEM GUI in the main STEM window, I made sure to check that the Clustering Method said, "STEM Clustering Method" and I did not change the defaults for the Maximum Number of Model Profiles or Maximum Unit Change in Model Profiles between Time Points.
9) I selected the yellow Execute button to run STEM. There were no errors and it ran successfully.
10) A new window opened that is called "All STEM Profiles (1)".
11) I clicked on a button that said "Interface Options...". There was a window that appeared below the text "X-axis scale should be:"
12) I clicked on the radio button that said "Based on real time".
13) I closed the interface options window.
14) I took a screenshot of the window by clicking the Alt and PrintScreen buttons at the same time.
15) I opened a powerpoint
16) I pasted the screenshot into powerpoint.
17) I clicked on each of the SIGNIFICANT profiles which are the colored images one at a time to show a more detailed plot that had all the genes for a specific profile.
18) I took screenshots of each of these and pasted them one by one into the powerpoint document I created.
19) I was supposed to then complete this step: * At the bottom of each profile window, there are two yellow buttons "Profile Gene Table" and "Profile GO Table". For each of the profiles, click on the "Profile Gene Table" button to see the list of genes belonging to the profile. In the window that appears, click on the "Save Table" button and save the file to your desktop. Make your filename descriptive of the contents, e.g. "wt_profile#_genelist.txt", where you replace the number symbol with the actual profile number.
I did not complete this step.
20) I added my journal entry to the wiki
21) I emailed Dr. Dahlquist about the missed step to see what I should do.
22) Uploading just the powerpoint for now since I am waiting to hear back about the other files.
23) Repeated the steps exactly for the STEM software. This time, I downloaded the Profile Gene Table and Profile GO Table.
24) I followed the labeling conventions... (Will check files to be sure)
25) I labeled my files so that they are clear and easy to know which file is which. I zipped them together. The file is named Andrew Tables Gene/Go Data.zip
26) The files in the zip are named dGLN3 #(whatever number they are) Go Table or Gene Table. We did not save one of these, and so I included a folder to remind that #45 Gene Table is missing. We used the output data from the table and the visual included in our powerpoint
Analysis of STEM Results
1) Select a profile of a gene you found interesting. Why?
I find profile 9 really interesting because I wonder why it went down in expression at first but then up in expression as it continued.
2) How many genes belong to this profile?
118 genes
3) How many genes were expected to belong to this profile?
34.1 genes were expected
4) What is the p value for the enrichment of genes in this profile?
1.3E -30 which is significant
5) I went to the GO table I saved. I selected the third row and chose Data> Filter> Autofilter. I filtered for p-values that are less than 0.05. There were 13 Go Terms associated with this profile with a p-value of <0.05
6) I then filtered for the corrected p-value column to show the corrected p-values under 0.05. There were 3 Go terms associated with this profile with a corrected p-value of <0.05.
7) I am going to look at GO:0055114, 0008652, 0008152, (these are the corrected p-values < 0.05), and GO: 0016787, 0005739, 0006629. I just chose
8) I went to the website [Gene Ontology] and copy and pasted each GO ID one at a time into the search field on the left of the screen. I would go to the the results page and click "Link to detailed information about (whatever specific go term was).
9)The term GO:0055114 came back as obsolete.
10) The term GO: 0008652 is amino acid biosynthetic process. The definition says "The chemical reactions and pathways resulting in the formation of amino acids, organic acids containing one or more amino substituents. Source: ISBN:0198506732" https://amigo.geneontology.org/amigo/term/GO:0008652
11) The term GO:0008152 is metabolic process. The definition says: "The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation. Source: ISBN:0198547684, GOC:go_curators" https://amigo.geneontology.org/amigo/term/GO:0008152
12) The term GO:0016787 came back as hydrolase activity. The definition came back as "Catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Source: ISBN:0198506732" https://amigo.geneontology.org/amigo/term/GO:0016787
13) The term GO:0005739 came back as mitochondrion. The definition was "A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration. Source: GOC:giardia, ISBN:0198506732" https://amigo.geneontology.org/amigo/term/GO:0005739
14) The term GO:0006629 came back as lipid metabolic process. The definition is "The chemical reactions and pathways involving lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. Includes fatty acids; neutral fats, other fatty-acid esters, and soaps; long-chain (fatty) alcohols and waxes; sphingoids and other long-chain bases; glycolipids, phospholipids and sphingolipids; and carotenes, polyprenols, sterols, terpenes and other isoprenoids. Source: GOC:ma" https://amigo.geneontology.org/amigo/term/GO:0006629
I think what is happening here is that the Gene profile is showing there is a drive to conserve energy and resources when the cold shock sets in especially given the processes involved here. Then is seems like maybe the cell is adapting to the cold and because of that it is able to start recovering which is why the metabolic process, the mitochondrion, the hydrolase activity, and metabolic processes are all increasing towards the end. I do wonder what is allowing the cell to adapt in the meantime. Im guessing it is probably some of the other clusters and the genes in those clusters.
Using YEASTRACT to infer which TF's regulate a cluster of genes
1) I opened the gene list for #9 and copied the gene ID's onto my clipboard.
2) I launched the browser and went to the YEASTRACT database by typing in www.yeastract.com. I did not click on the link because I was not signed in on the class website on the class computer.
3)I selected Rank by Tf and pasted my list of genes from my cluster into the box labeled ORFs/Genes.
4) I checked the box that said Check for all TFs and accepted the defaulted for the regulations filter (Documented, DNA binding or expression evidence)
5) I did not apply a filter for the "Filter Documented Regulations by environmental condition" option.
6) I ranked genes by TF using: The % of genes in the list and in YEASTRACT regulated by each TF.
7) I clicked on the Search button.
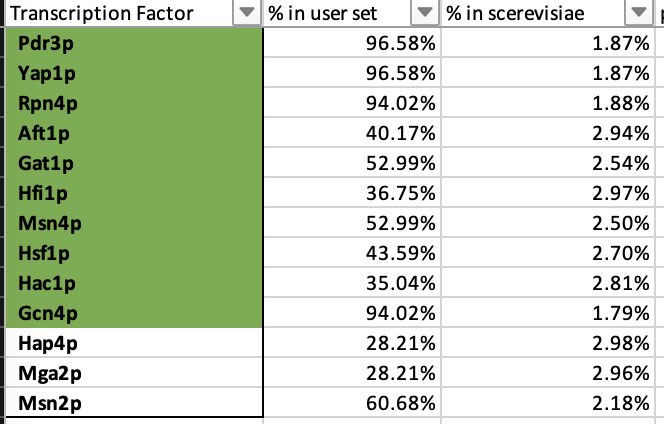
8) In the results window that appears, the p values colored green are considered "significant", the yellow ones are "borderline significant", and the pink ones are "not significant". How many TF's are green or significant?
9)I believe 10. The photo below shows my excel result. It may be 13. I am not sure. Unfortunately I did not copy down this number somewhere at the time thinking I would remember my methodology which I totally don't. I did highlight 10 green as you can see below.
10) CIN5 and GLN3 are both on the list. See Yellow Highlight for P value and % in YEASTRACT

GRNsight, creating and visualizing the GRN
1) I selected #9 from the list of significant transcription factors that I found in YEASTRACT
2) Navigated to the [GRNsight] beta website
3) Under the "Network" panel on the left-hand side, I clicked the button "Load from database".
4) I typed the standard name of each of the transcription factors in the "select gene" field one at a time. I would only enter one at which point I would click the find button which looks like a magnifying glass. Then I would enter the next TSP and hit find again, repeating the process.
5) I added 15 transcription factors
6) Not all of the rectangular boxes (nodes) were connected to another node. I went back to the load from database button and selected the transcription factors but this time I only selected the ones that were not disconnected.
7) The number of genes and edges in my network came out to 12 genes, 13 edges.
Creating the GRNmap Input Workbook
1) I exported my data from GRNsight to Excel. I did this by selecting the Export Menu and hitting "Export Data >To Excel". In the window that appeared there was an area that said "Select the Expression Data Source:". I chose the option "Dahlquist_2018"
2) Under "Select Workbook Sheets to Export:" I selected these: network dcin5_log2_expression dgln3_log2_expression wt_log2_expression degradation_rates optimization_parameters production_rates threshold_b
3) I clicked to "Export Workbook"
4) I opened the workbook to make sure it had all of these sheets listed. It did. None of the genes were missing a time block.
5) I checked the "production_rates" and the "degradation_rates" sheets and they had values for each gene.
6) The "threshold_b" sheet had a value of 0 for each gene.
7) I changed "alpha" in the "optimization_parameters" sheet to 0.02 from 0.002.
8) I inserted a new worksheet and named it "network_weights".
9) I copied the whole "network" sheet into the "network_weights" sheet.
10) I saved and added the file to this week 10 journal.
Data and Files
STEM PHOTOS FOR GLN3
Media:AS&CK_BIOL367_S24_STEM_PHOTOS_dGLN3.pptx
The Go Tables and Data Zipped
Media:Andrew&Charlotte_Tables_Gene-GoData.zip
The Text File for the STEM
The Results from YEASTRACT
Media:Asandle1YeastractResults9.xlsx
Photo of the GRNmap Results
Media:AndrewGRN(Yeastmine_-_SGD___2022-03-07;12_genes,_13_edges).png
The GRNmap exported and updated Excel
Media:GRN(Yeastmine_-_SGD_2022-03-07;12_genes,_13_edges)_weighted.xlsx
Acknowledgements & References
Acknowledgments
- I worked on this assignment with Charlotte Kaplan in class.
- We helped each other with the STEMs after not saving the data the first time.
- Dr.Dahlquist also helped us get back on track with this by meeting us before class.
- I also used some websites which are going to be listed in the resources, these websites were part of the instructions
- I also used the tools that were listed as tools in the methods like the STEM tool and GRNsight.
- Except for what is noted above, this individual journal entry was completed by me and not copied from another source.
References
- Dahlquist, K. Master_sheet_dGLN3.
- LMU BioDB 2024. (2024). Week 9. Retrieved Mar 20, 2024, from https://xmlpipedb.cs.lmu.edu/biodb/Spring2024/index.php/Week_9
- LMU BioDB 2024. (2024). Week 10. Retrieved Mar 30, 2024, from https://xmlpipedb.cs.lmu.edu/biodb/Spring2024/index.php/Week_10
- [GRNsight]
- [Gene Ontology]
- [Gene Ontology GO Term 0008652] Retrieved Apr 3, 2024
- [Gene Ontology GO Term 0008152] Retrieved Apr 3, 2024
- [Gene Ontology GO Term 0016787] Retrieved Apr 3, 2024
- [Gene Ontology GO Term 0005739] Retrieved Apr 3, 2024
- [Gene Ontology GO Term 0006629] Retrieved Apr 3, 2024
To User Page: User: Asandle1
To Template: Template:Asandle1