Difference between revisions of "Hivanson Week 10"
(→Data & Files: slides media added) |
(→References: fixed date) |
||
| Line 60: | Line 60: | ||
===Acknowledgments=== | ===Acknowledgments=== | ||
===References=== | ===References=== | ||
| − | *Help:Table. (2024). In Wikipedia. Retrieved | + | *Help:Table. (2024). In Wikipedia. Retrieved March 25, 2024, from https://en.wikipedia.org/w/index.php?title=Help:Table&oldid=1213890372 |
{{Template:Hivanson}} | {{Template:Hivanson}} | ||
Revision as of 18:24, 25 March 2024
Purpose
Methods/Results
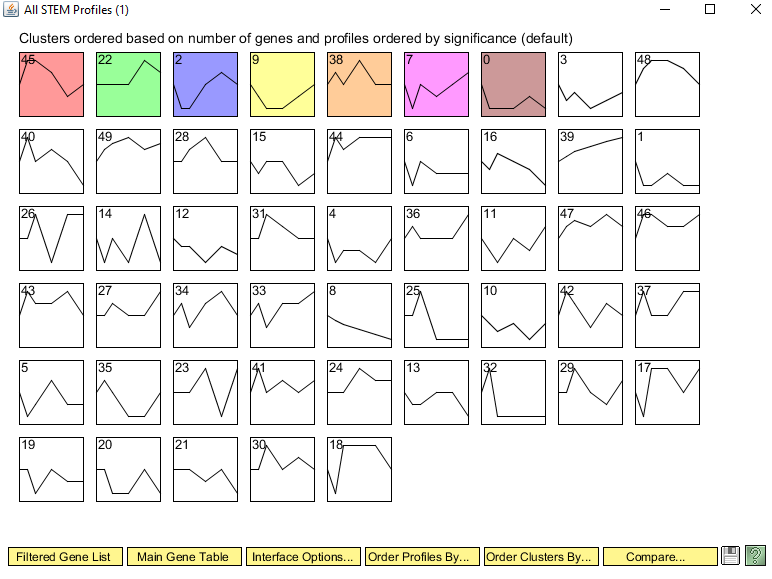
Why did you select this profile? In other words, why was it interesting to you? I selected profile 22 because the overall trend of a spike only at 90 minutes is different from the rest of the profiles' trends.
How many genes belong to this profile? 46 genes belong to profile 22.
How many genes were expected to belong to this profile? 23.6 genes were expected to be in profile 22.
What is the p value for the enrichment of genes in this profile? 2.3E-5
How many GO terms are associated with this profile at p < 0.05? 25
How many GO terms are associated with this profile with a corrected p value < 0.05? 7
| GO ID | Definition |
|---|---|
| GO:0034599 | cellular response to oxidative stress |
| GO:0005737 | cytoplasm |
| GO:0006897 | endocytosis |
| GO:0030479 | actin cortical patch |
| GO:0005856 | cytoskeleton |
| GO:0000324 | fungal-type vacuole |
Why does the cell react to cold shock by changing the expression of genes associated with these GO terms?
The genes that are associated with these GO terms include stress responses, like cellular response to oxidative stress, and its related terms that I excluded from the above table due to similarity. This makes sense as cold shock is considered a stressor. Additionally, various cellular structure components are associated with these GO terms, such as cytoplasm, cytoskeleton, and fungal-type vacuole which all may need to have regulated rigidity or some other physical characteristics at lower temperatures.
Also, what does this have to do with the transcription factor being deleted (for the groups working with deletion strain data) I want to compare our results to that of the wildtype. I don't understand completely what effects the CIN5 deletion should have, or what this does to the yeast. I tried reading the SGD description of CIN5. Will clarify with Dr. Dahlquist.
Data & Files
Excel file with microarray data for dCIN5
Tab text file with dCIN5 stem data
Stem dCIN5 significant profiles genelist folder
Stem dCIN5 significant profiles GOlist folder
Conclusion
Acknowledgments
References
- Help:Table. (2024). In Wikipedia. Retrieved March 25, 2024, from https://en.wikipedia.org/w/index.php?title=Help:Table&oldid=1213890372
- Hivanson
- Hivanson Week 1 | Week 1 Assignment
- Hivanson Week 2 | Week 2 Assignment
- IMD3 Hivanson and Nstojan1 Week 3 | Week 3 Assignment
- NeMO Week 4 | Week 4 Assignment
- Hivanson Week 5 | Week 5 Assignment
- Hivanson Week 6 | Week 6 Assignment
- Hivanson Week 8 | Week 8 Assignment
- Hivanson Week 9 | Week 9 Assignment
- Hivanson Week 10 | Week 10 Assignment
- Hivanson Week 12 | Week 12 Assignment
- Hivanson Week 13 | Week 13 Assignment
- Hivanson Week 14 | Week 14 Assignment
- Hivanson Week 15 | Week 15 Assignment
- Main page