Hivanson Week 10
Contents
Purpose
Methods/Results
Clustering and GO Term Enrichment with stem
- Preparing microarray data file for loading into STEM.
- I inserted a new worksheet into my Excel workbook, and named it "dCIN5_stem".
- I selected all of the data from the "ANOVA_dCIN5" worksheet and special pasted values into the "dCIN5_stem" worksheet.
- I renamed Column A to "SPOT" and Column B to "Gene Symbol." I deleted the column named "Standard_Name."
- I filtered the data in "dCIN5_B-H_p-value" > 0.05.
- Once the data has been filtered, I selected and deleted all of the rows (except for your header row). I undid the filter.
- I deleted all of the data columns except for the Average Log Fold change columns for timepoints 15m, 30m, 60m, 90m, and 120m.
- I renamed the data columns from "dCIN5_AvgLogFC_t
- I removed #DIV/0! errors using find and replace with nothing in the replace field.
- I saved the dCIN5_stem sheet as Text (Tab-delimited) (*.txt).
- I downloaded and extracted the STEM software. Click here to go to the STEM web site.
- I downloaded the Gene Ontology and yeast GO annotations from the 10 Wiki page.
- I launched the STEM program.
- Running STEM
- In section 1 (Expression Data Info) of the the main STEM interface window, I clicked on the Browse... button to navigate to and select the .txt file.
- I clicked on the radio button No normalization/add 0.
- I checked the box next to Spot IDs included in the data file.
- In section 2 (Gene Info) of the main STEM interface window, I left the default selection for the three drop-down menu selections for Gene Annotation Source, Cross Reference Source, and Gene Location Source as "User provided".
- I clicked the "Browse..." button to the right of the "Gene Annotation File" item. Browse to your "stem" folder and select the file "gene_association.sgd.gz" and click Open.
- In section 3 (Options) of the main STEM interface window, make sure that the Clustering Method says "STEM Clustering Method" and do not change the defaults for Maximum Number of Model Profiles or Maximum Unit Change in Model Profiles between Time Points.
- In section 4 (Execute) click on the yellow Execute button to run STEM.
- If you get an error, there are some known reasons why stem might not work. If you had #DIV/0! errors in your input file, it will cause problems. Re-open your file and open the Find/Replace dialog. Search for #DIV/0!, but don't put anything in the replace field. Click "Replace all" to remove the #DIV/0! errors. Then save your file and try again with stem.
- In section 1 (Expression Data Info) of the the main STEM interface window, I clicked on the Browse... button to navigate to and select the .txt file.
- Viewing and Saving STEM Results
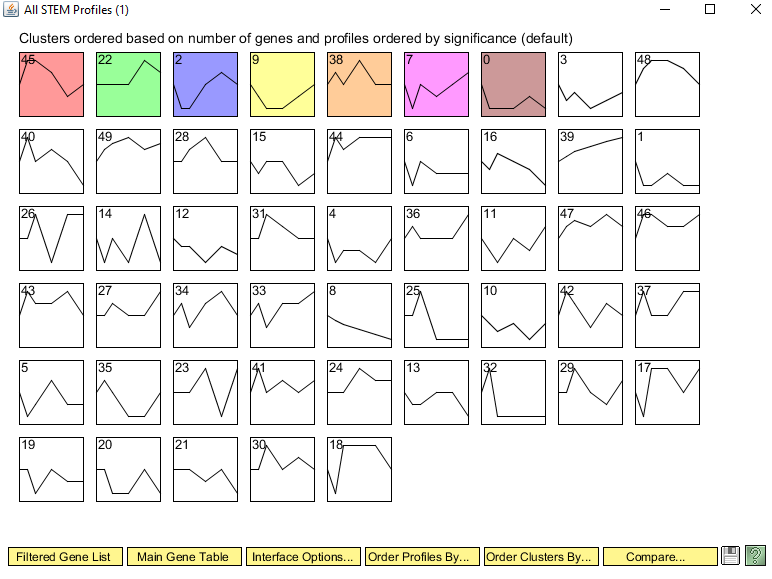
- A new window will open called "All STEM Profiles (1)". Each box corresponds to a model expression profile. Colored profiles have a statistically significant number of genes assigned; they are arranged in order from most to least significant p value. Profiles with the same color belong to the same cluster of profiles. The number in each box is simply an ID number for the profile.
- Click on the button that says "Interface Options...". At the bottom of the Interface Options window that appears below where it says "X-axis scale should be:", click on the radio button that says "Based on real time". Then close the Interface Options window.
- Take a screenshot of this window (on a PC, simultaneously press the
AltandPrintScreenbuttons to save the view in the active window to the clipboard) and paste it into a PowerPoint presentation to save your figures.
- Click on each of the SIGNIFICANT profiles (the colored ones) to open a window showing a more detailed plot containing all of the genes in that profile.
- Take a screenshot of each of the individual profile windows and save the images in your PowerPoint presentation.
- At the bottom of each profile window, there are two yellow buttons "Profile Gene Table" and "Profile GO Table". For each of the profiles, click on the "Profile Gene Table" button to see the list of genes belonging to the profile. In the window that appears, click on the "Save Table" button and save the file to your desktop. Make your filename descriptive of the contents, e.g. "wt_profile#_genelist.txt", where you replace the number symbol with the actual profile number.
- Upload these files to the wiki and link to them on your individual journal page. Note that it will be easier to zip all the files together and upload them as one file. To do this, select all of the files you want to zip together. Then right click and select "Send to" and "Compressed (zipped) folder" from the context menu.
- For each of the significant profiles, click on the "Profile GO Table" to see the list of Gene Ontology terms belonging to the profile. In the window that appears, click on the "Save Table" button and save the file to your desktop. Make your filename descriptive of the contents, e.g. "wt_profile#_GOlist.txt", where you use "wt", "dGLN3", etc. to indicate the dataset and where you replace the number symbol with the actual profile number. At this point you have saved all of the primary data from the STEM software and it's time to interpret the results!
- Upload these files to the wiki and link to them on your individual journal page. Note that it will be easier to zip all the files together and upload them as one file. To do this, select all of the files you want to zip together. Then right click and select "Send to" and "Compressed (zipped) folder" from the context menu.
- A new window will open called "All STEM Profiles (1)". Each box corresponds to a model expression profile. Colored profiles have a statistically significant number of genes assigned; they are arranged in order from most to least significant p value. Profiles with the same color belong to the same cluster of profiles. The number in each box is simply an ID number for the profile.
- Analyzing and Interpreting STEM Results
- Select one of the profiles you saved in the previous step for further intepretation of the data. I suggest that you choose one that has a pattern of up- or down-regulated genes at the cold shock timepoints. Each member of your group should choose a different profile. Answer the following:
- Why did you select this profile? In other words, why was it interesting to you?
- How many genes belong to this profile?
- How many genes were expected to belong to this profile?
- What is the p value for the enrichment of genes in this profile? Bear in mind that we just finished computing p values to determine whether each individual gene had a significant change in gene expression at each time point. This p value determines whether the number of genes that show this particular expression profile across the time points is significantly more than expected.
- Open the GO list file you saved for this profile in Excel. This list shows all of the Gene Ontology terms that are associated with genes that fit this profile. Select the third row and then choose from the menu Data > Filter > Autofilter. Filter on the "p-value" column to show only GO terms that have a p value of < 0.05. How many GO terms are associated with this profile at p < 0.05? The GO list also has a column called "Corrected p-value". This correction is needed because the software has performed thousands of significance tests. Filter on the "Corrected p-value" column to show only GO terms that have a corrected p value of < 0.05. How many GO terms are associated with this profile with a corrected p value < 0.05?
- Select 6 Gene Ontology terms from your filtered list (either p < 0.05 or corrected p < 0.05).
- Each member of the group will be reporting on his or her own cluster in your research presentation. You should take care to choose terms that are the most significant, but that are also not too redundant. For example, "RNA metabolism" and "RNA biosynthesis" are redundant with each other because they mean almost the same thing.
- Note whether the same GO terms are showing up in multiple clusters.
- Look up the definitions for each of the terms at http://geneontology.org. In your journal entry, will discuss the biological interpretation of these GO terms. In other words, why does the cell react to cold shock by changing the expression of genes associated with these GO terms? Also, what does this have to do with the transcription factor being deleted (for the groups working with deletion strain data)?
- To easily look up the definitions, go to http://geneontology.org.
- Copy and paste the GO ID (e.g. GO:0044848) into the search field on the left of the page.
- In the results page, click on the button that says "Link to detailed information about <term>, in this case "biological phase"".
- The definition will be on the next results page, e.g. here.
- Each member of the group will be reporting on his or her own cluster in your research presentation. You should take care to choose terms that are the most significant, but that are also not too redundant. For example, "RNA metabolism" and "RNA biosynthesis" are redundant with each other because they mean almost the same thing.
- Select one of the profiles you saved in the previous step for further intepretation of the data. I suggest that you choose one that has a pattern of up- or down-regulated genes at the cold shock timepoints. Each member of your group should choose a different profile. Answer the following:
Why did you select this profile? In other words, why was it interesting to you? I selected profile 22 because the overall trend of a spike only at 90 minutes is different from the rest of the profiles' trends.
How many genes belong to this profile? 46 genes belong to profile 22.
How many genes were expected to belong to this profile? 23.6 genes were expected to be in profile 22.
What is the p value for the enrichment of genes in this profile? 2.3E-5
How many GO terms are associated with this profile at p < 0.05? 25
How many GO terms are associated with this profile with a corrected p value < 0.05? 7
| GO ID | Definition |
|---|---|
| GO:0034599 | cellular response to oxidative stress |
| GO:0005737 | cytoplasm |
| GO:0006897 | endocytosis |
| GO:0030479 | actin cortical patch |
| GO:0005856 | cytoskeleton |
| GO:0000324 | fungal-type vacuole |
Why does the cell react to cold shock by changing the expression of genes associated with these GO terms?
The genes that are associated with these GO terms include stress responses, like cellular response to oxidative stress, and its related terms that I excluded from the above table due to similarity. This makes sense as cold shock is considered a stressor. Additionally, various cellular structure components are associated with these GO terms, such as cytoplasm, cytoskeleton, and fungal-type vacuole which all may need to have regulated rigidity or some other physical characteristics at lower temperatures.
Also, what does this have to do with the transcription factor being deleted (for the groups working with deletion strain data) I want to compare our results to that of the wildtype. I don't understand completely what effects the CIN5 deletion should have, or what this does to the yeast. I tried reading the SGD description of CIN5. Will clarify with Dr. Dahlquist.
Data & Files
Excel file with microarray data for dCIN5
Tab text file with dCIN5 stem data
Stem dCIN5 significant profiles genelist folder
Stem dCIN5 significant profiles GOlist folder
Conclusion
Acknowledgments
References
- Help:Table. (2024). In Wikipedia. Retrieved March 25, 2024, from https://en.wikipedia.org/w/index.php?title=Help:Table&oldid=1213890372
- Hivanson
- Hivanson Week 1 | Week 1 Assignment
- Hivanson Week 2 | Week 2 Assignment
- IMD3 Hivanson and Nstojan1 Week 3 | Week 3 Assignment
- NeMO Week 4 | Week 4 Assignment
- Hivanson Week 5 | Week 5 Assignment
- Hivanson Week 6 | Week 6 Assignment
- Hivanson Week 8 | Week 8 Assignment
- Hivanson Week 9 | Week 9 Assignment
- Hivanson Week 10 | Week 10 Assignment
- Hivanson Week 12 | Week 12 Assignment
- Hivanson Week 13 | Week 13 Assignment
- Hivanson Week 14 | Week 14 Assignment
- Hivanson Week 15 | Week 15 Assignment
- Main page