Asandle1 Week 13
To User Page: User: Asandle1 To Template: Template:Asandle1
Contents
Assignment Pages
Journals
Individual
Class Journals
Electronic Lab Notebook
Biological Databases P-val by Gene Initial Notes
Harbison Paper
To get to the Team Journal Page please click here: Yeast Beasts
Things I notice
- The document is called Pval by Gene which makes me think that we are looking at the Pvalues in the genes which tracks considering what the paper is based on.
- The document has 6,231 rows which aligns with the gene numbers from the paper
- The document has 206 columns, 203 are values. This makes me think the columns are for each transcriptional regulator.
- There are some value boxes that just say NaN. Not sure how to deal with these
- We have the gene names in column 1
- We have the gene tags that humans can actually read in column 2
- We have plain english descriptions of what everything is in column 3
Thought Process:
Layout Assumptions: (Maybe incorrect)
- Primary Key should be Gene ID Column for the Data
Questions:
- Data preprocessing?
- Can we worry about the NaN entries once everything has been added to Access or do we need to figure out how to remove those without removing every entry?
- How do we want to import by?
- We want to be able to view by Gene or by environmental difference, does this mean making an access entry for the Genes and for the different experimental conditions?
- How do we make sure the database for the Harbison paper also works with all the others? I think this probably has to do with the primary key which then means we can really only organize across the Gene ID’s because that is what will be common across other experiments.
- Is there anything I am missing?
In Class Tuesday April 16th Notes
Added a Folder to the BIOL367_Spring2024 Box Folder and Labeled it Dean and Andrew Database File Folder
If there is a significant P value we make it a 1 anything else we give a 0. We will do =IF(pval<0.01,1,0)
Made a copy of YPD and called it YPD edits so I have the original saved in case of any errors.
I then did =if(YPD!D3 < 0.001,1,0) in my new copy document referring to the same cell in the original document.
I dragged my selection across the top line of P-values and then double clicked to expand down the entire data selection.
I then filtered the data in column D and selected only the check box for 1's. It came back with 6 results which was the same number of results that Dr. Dahlquist found.
Then I cleared the filter.
I removed the top row that had numbers that I didn't have any context for.
Next I removed the _YPD at the end of the row that was now the new top row. I replaced it with nothing, using the find and replace feature and selected replace all.
Then I filtered again to double check all my data and formulas still existed and worked correctly and it did.
File:Pvalbygene forpaper abbrAndrewDay1.xls
After Class
Take a look at this image:
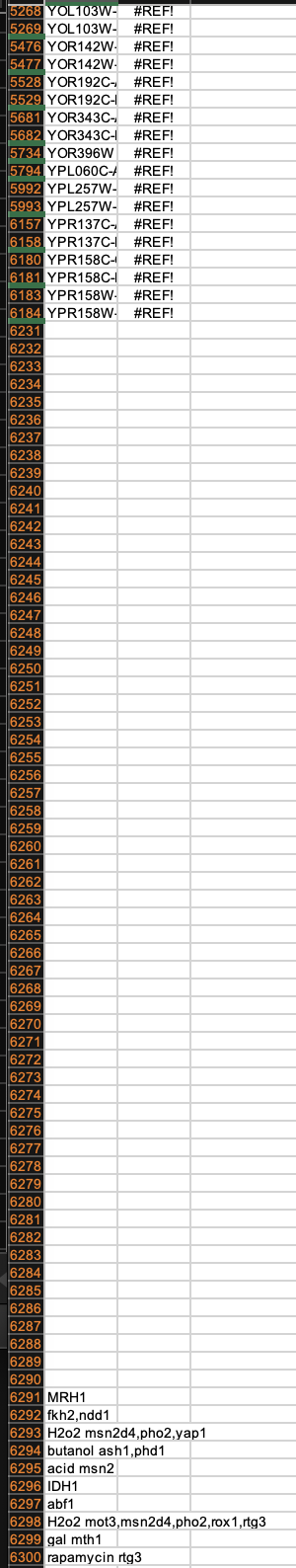
![]() You can see that the shorthand name and the description are missing in line 37. This happens in other places as well.
You can see that the shorthand name and the description are missing in line 37. This happens in other places as well.
For now, I am going to sort by blanks and ones that have Ref in them so I can find what needs fixing
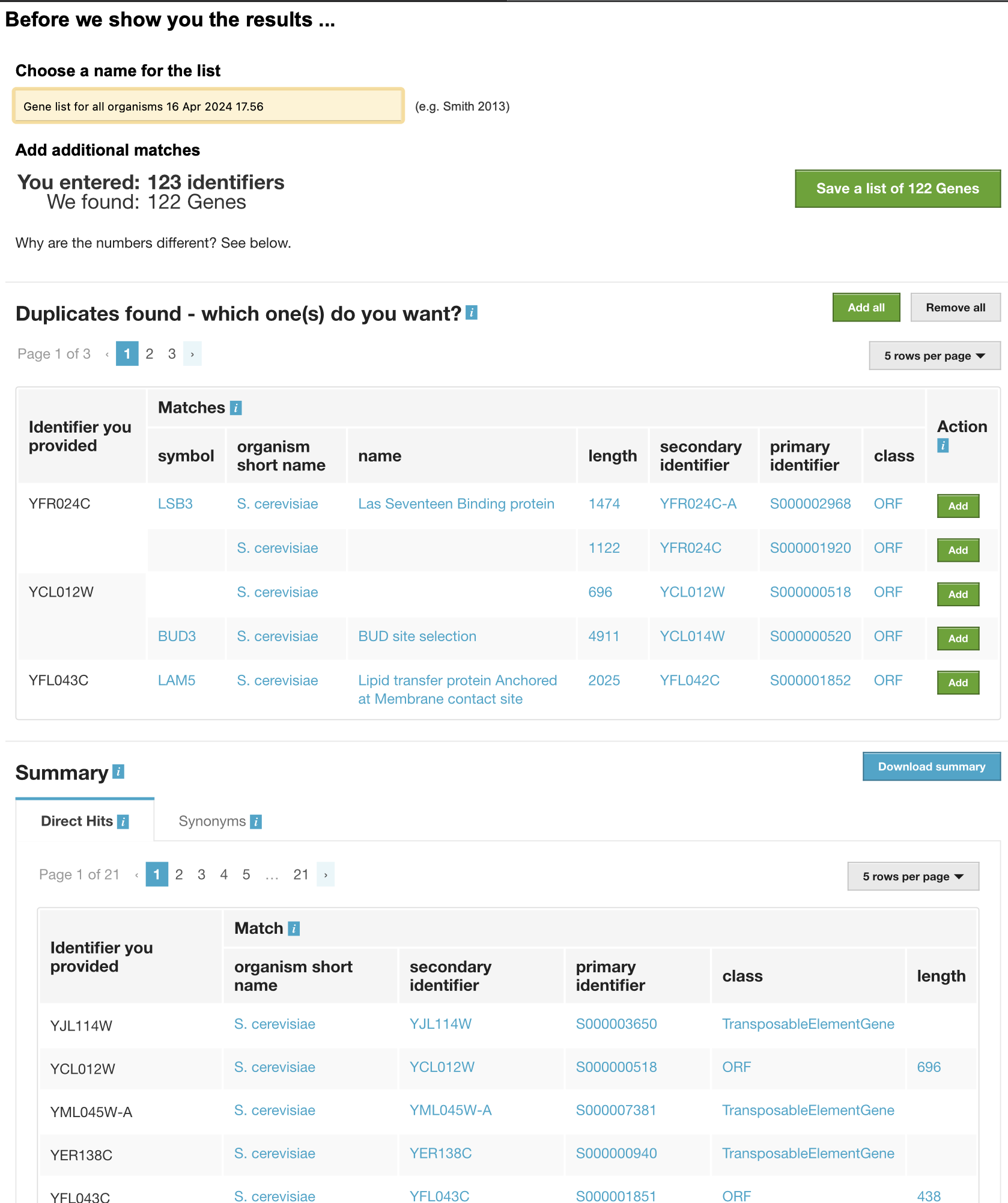
Now I noticed after filtering another issue, which the image below shows. (Blanks and then some random information that I am unsure about.)
I tried to go to Yeastmine with the list of identifiers for the missing symbols but realized this would be a really slow process and there is probably a better way. Also wasn't sure which to select. Here is what I saw.
References & Acknowledgements
- Dr. Dahlquist helped Dean and I with the excel file steps, specifically relating to setting up the =if equation to change the significant P-values to 1 and the non-significant values to 0. This was in class on tuesday. She also responded to some questions I had asked her over email that can be found at the top of this journal.
- I spoke with Dean and uploaded my excel file since he was having difficulties. I also spoke to him Wednesday April 17th at around 9 P.M. over text.
- LMU BioDB 2024. (2024). Coder/Designer. Retrieved April 17, 2024, from https://xmlpipedb.cs.lmu.edu/biodb/spring2024/index.php/Coder/Designer
- Community Central Fandom. (2024). Forum:How do I link to a heading on another page?. Retrieved April 17, 2024, from https://community.fandom.com/wiki/Forum:How_do_I_link_to_a_heading_on_another_page%3F
Except for what is noted above, this individual journal entry was completed by me and not copied from another source. Asandle1 (talk)