Difference between revisions of "CMR2/YOR093C Week 3"
(addeed breaks) |
(→What was different about the information provided about your gene in each of the parent databases?: revised paragraph) |
||
| (38 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
== Individual Journal Assignment == | == Individual Journal Assignment == | ||
| − | + | === Summary === | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | The CMR2 gene found in ''Saccharomyces cerevisiae'' is a unique gene having a longer lifespan. Not much is studied on the gene itself, but it is known from literature that this gene can live for as long as five years. Found within the cytosol of Saccharomyces cerevisiae cells, this gene is able to withstand certain harmful chemicals and low temperatures to keep itself alive. As said previously, this gene is relatively new in its discovery, leading to its lack of highly informational literature on it's full functions, however, the studies that have been carried out have shown that, while still being relatively viable, yeast cells with the CMR2 a.k.a. YORO93 gene are in some way less competitively fit than those without the activated gene, however, when the gene is deleted from the genome, the mutant becomes sensitive to factors that create responses for unfolded protein. This gene is full of many secrets, but hopefully as time goes on, more studies on the gene will allow for great discoveries on its functions and capabilities [https://www.yeastgenome.org/locus/S000005619#reference As show by the SGD database]. | |
| − | + | *'''Standard Name:''' CMR2 | |
| + | *'''Systematic Name:''' YOR093C | ||
| + | *'''Name Description:''' Changed Mutation Rate | ||
| + | *'''Gene ID:''' | ||
| + | ** [https://www.yeastgenome.org/locus/S000005619#overview SGD]: S000005619 | ||
| + | ** [https://www.ncbi.nlm.nih.gov/gene/854260 NCBI]: 854260 | ||
| + | ** [https://uswest.ensembl.org/Saccharomyces_cerevisiae/Gene/Summary?db=core;g=YOR093C;r=XV:497506-502452;t=YOR093C_mRNA Ensembl]: R64-1-1:BK006948.2 | ||
| + | ** [https://www.uniprot.org/uniprot/Q12275 UniPort]: 559292 | ||
| − | === | + | === What is the DNA sequence of your gene? === |
| − | '' | + | 1 ATGGATTTTT CTATTCCTCC TACCTTACCT CTAGATCTGC AAAGTCGGTT AAATGAGTTA <br> |
| + | 61 ATTCAAGACT ATAAGGATGA AAATCTAACA AGAAAGGGTT ATGAGACCAA AAGAAAACAA <br> | ||
| + | 121 TTGCTAGACA AATTTGAAAT CTCCCAAATG AGGCCTTATA CCCCGCTTCG ATCTCCCAAT <br> | ||
| + | 181 TCGAGAAAGT CGAAACATTT GCACAGAAGA AATACAAGTC TTGCATCATC CATTACGTCG <br> | ||
| + | 241 TTACCTAATT CAATTGACAG ACGTCATTCT ATCTATCGTG TCACAACTAT AAACTCGACT <br> | ||
| + | 301 TCTGCGAACA ATACTCCCAG AAGGCGGAGC AAGAGATATA CAGCTTCTTT GCAATCGTCA <br> | ||
| + | 361 TTGCCAGGTT CCAGTGATGA AAACGGTTCA GTGAAGGATG CCGTTTATAA TCCTATGATA <br> | ||
| + | 421 CCATTACTAC CAAGACACAC TGGAGCCGAA AATACTAGCA GTGGAGACTC AGCAATGACG <br> | ||
| + | 481 GATTCTTTAC CGCTAATTTT ACGGGGACGC TTTGAACATT ATGATGGTCA AACAGCGATG <br> | ||
| + | 541 ATCAGCATAA ATTCTAAAGG CAAAGAAACG TTCATCACAT GGGATAAACT TTATTTGAAA <br> | ||
| + | 601 GCGGAGAGGG TAGCACATGA ATTGAATAAG AGTCACCTTT ACAAAATGGA TAAGATTTTA <br> | ||
| + | 661 CTATGGTACA ATAAAAATGA TGTCATCGAA TTTACTATTG CATTATTAGG CTGTTTCATA <br> | ||
| + | 721 TCCGGCATGG CAGCTGTACC AGTCTCATTC GAAACGTATT CTCTGCGTGA GATCCTTGAG <br> | ||
| + | 781 ATAATTAAAG TAACAAATTC GAAATTTGTT CTGATTTCTA ACGCATGTCA TAGACAATTA <br> | ||
| + | 841 GATAACTTGT ATTCGTCATC AAATCATTCA AAAGTTAAAC TGGTTAAGAA CGATGTTTTC <br> | ||
| + | 901 CAGCAAATCA AATTTGTCAA GACTGATGAC TTAGGGACGT ACACAAAGGC AAAAAAAACT <br> | ||
| + | 961 TCGCCTACCT TTGATATACC GAATATTTCA TATATAGAGT TTACTAGAAC ACCGCTAGGT <br> | ||
| + | 1021 CGTCTTTCTG GCGTCGTCAT GAAGCATAAC ATCTTAATAA ACCAATTTGA AACGATGACA <br> | ||
| + | 1081 AAAATTCTAA ACTCACGATC AATGCCTCAC TGGAAACAAA AATCACAAAG CATCAGGAAG <br> | ||
| + | 1141 CCATTTCACA AAAAAATTAT GGCAACCAAC TCAAGGTTTG TTATCTTGAA TAGTCTGGAC <br> | ||
| + | 1201 CCTACTAGAT CTACTGGTTT GATAATGGGT GTGCTTTTCA ATCTTTTTAC TGGTAATCTT <br> | ||
| + | 1261 ATGATATCTA TCGATAGTAG TATATTACAA CGACCTGGGG GCTATGAAAA TATCATTGAT <br> | ||
| + | 1321 AAATTTAGAG CCGACATTTT ACTGAATGAT CAGCTACAAT TGAAGCAGGT AGTTATTAAT <br> | ||
| + | 1381 TATTTGGAAA ATCCCGAATC AGCTTTTTCC AAGAAGCACA AAATAGATTT CAGTTGCATC <br> | ||
| + | 1441 AAGTCATGTT TAACCTCATG TACCACTATA GATACCGATG TAAGCGAGAT GGTTGTTCAT <br> | ||
| + | 1501 AAATGGCTGA AAAATTTAGG ATGTATTGAT GCACCATTTT GTTATTCTCC CATGTTAACG <br> | ||
| + | 1561 TTATTGGACT TTGGTGGTAT CTTTATATCT ATAAGAGATC AATTAGGAAA CCTAGAGAAC <br> | ||
| + | 1621 TTTCCAATAC ATAATTCAAA ACTAAGGCTT CAAAATGAAT TGTTTATAAA CCGCGAGAAA <br> | ||
| + | 1681 TTGAAGCTTA ATGAGGTTGA ATGTAGCATA ACTGCTATGA TTAATTCATC AAGCTCCTTC <br> | ||
| + | 1741 AAAGATTATC TTAAATTAGA AACATTTGGC TTCCCAATTC CTGATATCAC TTTATGTGTG <br> | ||
| + | 1801 GTTAATCCAG ATACGAATAC GTTGGTGCAA GACCTCACTG TTGGAGAAAT ATGGATCTCT <br> | ||
| + | 1861 TCTAACCACA TCACCGACGA ATTTTACCAG ATGGATAAAG TGAACGAATT TGTGTTCAAA <br> | ||
| + | 1921 GCAAAACTAA ATTACTCCGA AATGTTTTCT TGGGCAAAGT ATGAAATGCC TACTAACGAA <br> | ||
| + | 1981 AAATCGCAAG CAGTTACTGA ACAATTGGAT ACCATTCTGA ATATTTGTCC TGCAAATACC <br> | ||
| + | 2041 TACTTTATGA GAACCAAGCT TATGGGGTTT GTTCATAACG GAAAGATATA CGTGCTTTCA <br> | ||
| + | 2101 CTCATAGAAG ATATGTTTTT ACAGAATAGA TTAATCAGAT TACCTAACTG GGCTCACACA <br> | ||
| + | 2161 TCAAACCTTC TTTATGCCAA AAAAGGGAAT CAATCTGCTC AACCTAAAGG TAACACTGGA <br> | ||
| + | 2221 GCAGAAAGCA CTAAGGCTAT TGATATTTCG AGTTTAAGTG GCGAAACATC TTCGGGATAC <br> | ||
| + | 2281 AAAAGAGTTG TTGAATCCCA CTATCTACAA CAGATTACTG AAACAGTTGT GAGGACAGTA <br> | ||
| + | 2341 AATACAGTTT TTGAGGTTGC TGCATTTGAA TTACAACACC ATAAAGAAGA GCACTTTTTA <br> | ||
| + | 2401 GTTATGGTGG TAGAAAGCTC CCTAGCAAAA ACCGAAGAAG AGAGTAAAAA TGGCGAAACA <br> | ||
| + | 2461 ACAGATACTA CCTTAATGAA ATTTGCAGAA ACACAGAGGA ACAAATTAGA AACGAAAATG <br> | ||
| + | 2521 AATGATTTAA CTGACCAAAT TTTTAGAATT CTTTGGATTT TTCATAAAAT CCAACCAATG <br> | ||
| + | 2581 TGCATTTTAG TTGTGCCGAG AGATACTTTA CCTAGGAGAT ATTGTTCTCT GGAATTGGCC <br> | ||
| + | 2641 AATAGCACGG TAGAGAAGAA GTTTTTAAAC AATGATCTCA GTGCACAGTT TGTAAAGTTT <br> | ||
| + | 2701 CAATTCGATA ATGTCATATT GGATTTTTTG CCCCATTCAG CGTACTATAA TGAGAGTATA <br> | ||
| + | 2761 TTATCCGAGC ATTTATCAAA ATTGAGAAAG ATGGCTTTAC AAGAAGAATA CGCTATGATC <br> | ||
| + | 2821 GAACCAGCAT ATCGTAATGG CGGTCCCGTT AAACCAAAAC TTGCCCTACA ATGCAGTGGT <br> | ||
| + | 2881 GTTGATTACA GAGATGAGTC TGTTGACACT CGAAGCCATA CAAAACTCAC AGACTTCAAA <br> | ||
| + | 2941 TCGATTTTAG AGATTCTGGA ATGGAGAATT TCTAATTATG GGAACGAAAC GGCGTTCAGT <br> | ||
| + | 3001 GATGGTACAA ACACAAACCT AGTCAACTCT TCAGCTAGTA ACGATAATAA CGTTCACAAA <br> | ||
| + | 3061 AAGGTATCAT GGGCAAGTTT TGGTAAAATT GTCGCAGGCT TTTTGAAAAA AATCGTAGGT <br> | ||
| + | 3121 TCTAAAATTC CATTGAAGCA TGGTGACCCT ATTATTATTA TGTGTGAAAA TTCGGTTGAA <br> | ||
| + | 3181 TACGTAGCGA TGATCATGGC CTGCCTGTAC TGTAACTTAT TGGTAATTCC CCTACCAAGC <br> | ||
| + | 3241 GTTAAGGAGT CTGTCATAGA AGAGGACCTA AAAGGCTTGG TTAATATTAT TCAAAGTTAC <br> | ||
| + | 3301 AAAGTGAAAA GAGTATTTGT TGATGCTAAA TTGCACTCAT TGTTGAATGA TAATAATGTC <br> | ||
| + | 3361 GTAAACAAAT GTTTTAAGAA ATACAAAAGT TTGATACCCA AGATTACAGT TTTCTCAAAA <br> | ||
| + | 3421 GTCAAGACAA AGAATGCGTT AACAGTATCT ATGTTTAAGA ATGTGTTAAA GCAGAAATTT <br> | ||
| + | 3481 GGGGCCAAAC CAGGTACTAG GATTGGTATG ACACCGTGTG TTGTATGGGT AAATACAGAG <br> | ||
| + | 3541 TATGATGTAA CATCTAATAT TCACGTAACA ATGACACATT CGTCTTTACT GAATGCAAGC <br> | ||
| + | 3601 AAAATAGTCA AAGAAACTTT GCAGTTAAGA AATAATAGTC CACTTTTCTC TATATGTTCT <br> | ||
| + | 3661 CATACATCTG GATTGGGCTT TATGTTCAGC TGTTTGTTAG GAATTTATAC GGGTGCTTCA <br> | ||
| + | 3721 ACTTGCTTAT TCAGCCTTAC TGATGTTCTT ACTGACCCTA AGGAGTTTTT AATTGGCCTT <br> | ||
| + | 3781 CAAAATTTAA ACGTAAAAGA TTTATATTTG AAGCTTGAAA CGTTATATGC TTTACTGGAT <br> | ||
| + | 3841 AGAGCCTCTA GTTTGATTGA GGGATTCAAA AATAAGAAGG AGAATATAAA CTCTGCCAAA <br> | ||
| + | 3901 AACAATACAT CTGGCTCGCT TAGAGAAGAT GTTTTCAAGG GTGTTCGAAA TATCATGATA <br> | ||
| + | 3961 CCTTTTCCTA ATAGACCAAG GATTTATACA ATTGAAAATA TTTTAAAACG GTACTCAACC <br> | ||
| + | 4021 ATATCCCTAT CATGTACACA AATAAGTTAT GTCTATCAAC ATCACTTCAA TCCGCTTATA <br> | ||
| + | 4081 TCATTAAGGT CGTATCTGGA TATTCCTCCG GTTGACCTAT ATTTAGACCC ATTTTCACTG <br> | ||
| + | 4141 AGGGAAGGTA TAATTAGAGA AGTCAACCCC AATGATGTAA GTGCTGGAAA CTATATCAAG <br> | ||
| + | 4201 ATTCAAGATT CTGGTGTTGT TCCTGTATGT ACTGACGTTT CAGTAGTTAA TCCAGAGACA <br> | ||
| + | 4261 CTCCTACCAT GTGTTGATGG AGAATTCGGT GAGATTTGGT GCTGTTCGGA AGCTAATGCA <br> | ||
| + | 4321 TTCGACTACT TTGTGTGTAA CAGTTCAAAA AACAAGTTGT ACAAAGATCC CTTTATCACA <br> | ||
| + | 4381 GAACAATTCA AAAGTAAGAT GAAGAGCGAA GTGAATAACA CTTTAAGCTA TTTGAGAACT <br> | ||
| + | 4441 GGTGATCTGG GCTTTATCAA AAACGTAAGT TGCACGAATT CACAGGGAGA GGTCGTCAAT <br> | ||
| + | 4501 TTGAACTTGT TGTTTGTTTT GGGAAGTATT CATGAATCCA TTGAAATTCT AGGATTGACA <br> | ||
| + | 4561 CATTTTGTTA GTGATTTGGA GAGAACCGTT AAAGATGTTC ACAGTGACAT TGGTAGTTGT <br> | ||
| + | 4621 TTGATTGCCA AAGCAGGTGG ATTATTAGTG TGCTTAATTA GGTGTAAGGA ACGACATAAT <br> | ||
| + | 4681 CCCATCTTGG GTAATCTAAC GACTTTAATT GTTTCAGAGT TGTTAAATAA GCATGGTGTC <br> | ||
| + | 4741 ATTTTAGATT TATGCACATT TGTGAGAACT AAAGGTATAA GCCCAAAAAA TTCTAGTATG <br> | ||
| + | 4801 ATAATGGAAG TTTGGGCGAA AAATAGAGCA TCGATAATGC AAGCCTGGTT TGATCAGAAA <br> | ||
| + | 4861 ATTCAAATAG AAGCACAATT TGGCATAAAC TATGGTGAAA ATATTTCCAT TTATTTATTA <br> | ||
| + | 4921 TCAGATTATG AAAAGGACAA TATTTAA<br> | ||
| + | ''Sequence Acquired from SGD'' | ||
| − | + | === What is the protein sequence corresponding to your gene? === | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
1 MDFSIPPTLP LDLQSRLNEL IQDYKDENLT RKGYETKRKQ LLDKFEISQM RPYTPLRSPN <br> | 1 MDFSIPPTLP LDLQSRLNEL IQDYKDENLT RKGYETKRKQ LLDKFEISQM RPYTPLRSPN <br> | ||
| Line 155: | Line 131: | ||
1561 PILGNLTTLI VSELLNKHGV ILDLCTFVRT KGISPKNSSM IMEVWAKNRA SIMQAWFDQK <br> | 1561 PILGNLTTLI VSELLNKHGV ILDLCTFVRT KGISPKNSSM IMEVWAKNRA SIMQAWFDQK <br> | ||
1621 IQIEAQFGIN YGENISIYLL SDYEKDNI*<br> | 1621 IQIEAQFGIN YGENISIYLL SDYEKDNI*<br> | ||
| + | ''Sequence Acquired from SGD'' | ||
| + | |||
| + | [[Image:CMR2_Protein_Sequence.png | thumb | Protein Sequence of CMR2 in the first reading frame, Reading Frame #1]] | ||
| + | |||
| + | === What is the function of your gene? === | ||
| + | *CMR2 functions include: Extending the lifespan of yeast bacteria, increase chemical resistance, and increase catalytic activity. ''Question answered with SGD'' | ||
| + | === What was different about the information provided about your gene in each of the parent databases? === | ||
| + | * Each database consists of similar information relating to CMR2. In regards to content, each database has similar information about the functions of the gene and the molecular aspects of it, but the gene identifiers are different. Each database has their own unique identifier. In addition, Ensembl also references Uniprot with the identifier name and provides a link to more information on the Uniprot page. Each database also has different research articles that tested the gene. The literature had similar concepts, but the experiments were performed by completely different scientists that may have referenced each other when completing it. In regards to the presentation of the database, NCBI and Ensembl were more complex to navigate in comparison to SGD and Uniprot. NCBI and Ensembl had more of the molecular level type information including interactions that the gene had with other genes, and had more navigation links that connected the interactions with the genes. SGD and Uniprot provided more information on the performance of the gene in a yeast cell as well as the phenotype data from it including its resistance to chemicals. The information in SGD and Uniprot were more organized and had different categories and subsections that made it easier to navigate. ''Question answered with SGD, Ensembl, Uniprot, and NCBI'' | ||
| + | |||
| + | === Why did you choose your particular gene? i.e., why is it interesting to you and your partner? === | ||
| + | *We chose the CMR2 gene because we were interested in learning about a gene that had a longer lifespan than others. In a study done on the CMR2 gene in ''Saccharomyces cerevisiae'', Postma and other scientists found out that cells containing this gene along with a few other genes, were able to survive for about 5 years on YPD plates due to their ability to undergo long periods of low metabolic rates (Postma et al., 2009). From this paper, we were curious to research the different ways that scientists would stress this gene enough to cause an apoptotic cell death. The literature has been consistent with their research indicating that the presence of this gene in ''Saccharomyces cerevisiae'' will decrease cell death. ''Question answered with SGD'' | ||
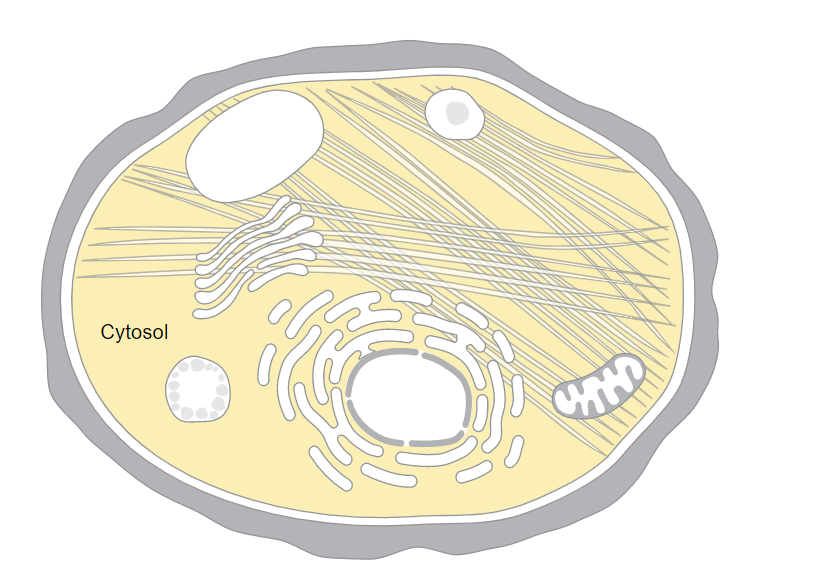
| + | [[File:CMR2.PNG | thumb | Location of gene within a yeast cell. The locations include presence in the cytosol, cellular bud neck, and the integral component of the membrane]] | ||
| + | |||
| + | === Acknowledgements === | ||
| + | * We would like to thank [[User:Kdahlquist| Dr. Kam Dahlquist]] for the introduction to the databases utilized during this research assignment. | ||
| + | * Thank you to my homework partner [[User:Dmadere| Delisa Madere]] for her partnership in this assignment. We met in person and spoke over the phone in order to discuss the sharing of responsibility, contents of the page, and understanding of the assignment. All work was shared between the two of us. | ||
| + | *Except for what is noted above, this journal entry was done by me and not copied from another source. [[User:Jnimmers|Jnimmers]] ([[User talk:Jnimmers|talk]]) 17:29, 18 September 2019 (PDT) | ||
| + | |||
| + | * I worked with my homework partner [[User:Jnimmers|Joey Nimmers]] on this assignment this week both in class and through cellular communication. We split the work evenly between us and contributed to all aspects of the project. | ||
| + | * We consulted the Wiki Help page when designing the format of the assignment as well. [[http://mediawiki.org/wiki/Help:Contents Help Page]] | ||
| + | *'''"Except for what is noted above, this individual journal entry was completed by me and not copied from another source."''' [[User:Dmadere|Dmadere]] ([[User talk:Dmadere|talk]]) 22:25, 18 September 2019 (PDT) | ||
| + | |||
| + | === References === | ||
| + | *The Ensembl Project. (n.d.). Retrieved from https://uswest.ensembl.org/Saccharomyces_cerevisiae/Info/Index | ||
| + | |||
| + | *UniProt Consortium European Bioinformatics Institute Protein Information Resource SIB Swiss Institute of Bioinformatics. (n.d.). UniProt Consortium. Retrieved from https://www.uniprot.org/ | ||
| + | |||
| + | *Saccharomyces Genome Database | SGD. (n.d.). Retrieved from https://www.yeastgenome.org/ | ||
| + | |||
| + | *Home - Gene - NCBI. (n.d.). Retrieved from https://www.ncbi.nlm.nih.gov/gene | ||
| + | |||
| + | *Postma, L., Lehrach, H., & Ralser, M. (2009). Surviving in the cold: yeast mutants with extended hibernating lifespan are oxidant sensitive. Aging, 1(11), 957–960. doi:10.18632/aging.100104 | ||
| + | |||
| + | *Teng, X., Cheng, W. C., Qi, B., Yu, T. X., Ramachandran, K., Boersma, M. D., … Hardwick, J. M. (2011). Gene-dependent cell death in yeast. Cell death & disease, 2(8), e188. doi:10.1038/cddis.2011.72 | ||
| + | *Wiki page week 3 assignment updates as listed by LMU BioDB 2019. (2019). Week 1. Retrieved September 18, 2019, from https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/Week_1 | ||
| − | + | {{Dmadere}} | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | {{Jnimmers}} | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Latest revision as of 21:54, 18 September 2019
Contents
- 1 Individual Journal Assignment
- 1.1 Summary
- 1.2 What is the DNA sequence of your gene?
- 1.3 What is the protein sequence corresponding to your gene?
- 1.4 What is the function of your gene?
- 1.5 What was different about the information provided about your gene in each of the parent databases?
- 1.6 Why did you choose your particular gene? i.e., why is it interesting to you and your partner?
- 1.7 Acknowledgements
- 1.8 References
Individual Journal Assignment
Summary
The CMR2 gene found in Saccharomyces cerevisiae is a unique gene having a longer lifespan. Not much is studied on the gene itself, but it is known from literature that this gene can live for as long as five years. Found within the cytosol of Saccharomyces cerevisiae cells, this gene is able to withstand certain harmful chemicals and low temperatures to keep itself alive. As said previously, this gene is relatively new in its discovery, leading to its lack of highly informational literature on it's full functions, however, the studies that have been carried out have shown that, while still being relatively viable, yeast cells with the CMR2 a.k.a. YORO93 gene are in some way less competitively fit than those without the activated gene, however, when the gene is deleted from the genome, the mutant becomes sensitive to factors that create responses for unfolded protein. This gene is full of many secrets, but hopefully as time goes on, more studies on the gene will allow for great discoveries on its functions and capabilities As show by the SGD database.
- Standard Name: CMR2
- Systematic Name: YOR093C
- Name Description: Changed Mutation Rate
- Gene ID:
What is the DNA sequence of your gene?
1 ATGGATTTTT CTATTCCTCC TACCTTACCT CTAGATCTGC AAAGTCGGTT AAATGAGTTA
61 ATTCAAGACT ATAAGGATGA AAATCTAACA AGAAAGGGTT ATGAGACCAA AAGAAAACAA
121 TTGCTAGACA AATTTGAAAT CTCCCAAATG AGGCCTTATA CCCCGCTTCG ATCTCCCAAT
181 TCGAGAAAGT CGAAACATTT GCACAGAAGA AATACAAGTC TTGCATCATC CATTACGTCG
241 TTACCTAATT CAATTGACAG ACGTCATTCT ATCTATCGTG TCACAACTAT AAACTCGACT
301 TCTGCGAACA ATACTCCCAG AAGGCGGAGC AAGAGATATA CAGCTTCTTT GCAATCGTCA
361 TTGCCAGGTT CCAGTGATGA AAACGGTTCA GTGAAGGATG CCGTTTATAA TCCTATGATA
421 CCATTACTAC CAAGACACAC TGGAGCCGAA AATACTAGCA GTGGAGACTC AGCAATGACG
481 GATTCTTTAC CGCTAATTTT ACGGGGACGC TTTGAACATT ATGATGGTCA AACAGCGATG
541 ATCAGCATAA ATTCTAAAGG CAAAGAAACG TTCATCACAT GGGATAAACT TTATTTGAAA
601 GCGGAGAGGG TAGCACATGA ATTGAATAAG AGTCACCTTT ACAAAATGGA TAAGATTTTA
661 CTATGGTACA ATAAAAATGA TGTCATCGAA TTTACTATTG CATTATTAGG CTGTTTCATA
721 TCCGGCATGG CAGCTGTACC AGTCTCATTC GAAACGTATT CTCTGCGTGA GATCCTTGAG
781 ATAATTAAAG TAACAAATTC GAAATTTGTT CTGATTTCTA ACGCATGTCA TAGACAATTA
841 GATAACTTGT ATTCGTCATC AAATCATTCA AAAGTTAAAC TGGTTAAGAA CGATGTTTTC
901 CAGCAAATCA AATTTGTCAA GACTGATGAC TTAGGGACGT ACACAAAGGC AAAAAAAACT
961 TCGCCTACCT TTGATATACC GAATATTTCA TATATAGAGT TTACTAGAAC ACCGCTAGGT
1021 CGTCTTTCTG GCGTCGTCAT GAAGCATAAC ATCTTAATAA ACCAATTTGA AACGATGACA
1081 AAAATTCTAA ACTCACGATC AATGCCTCAC TGGAAACAAA AATCACAAAG CATCAGGAAG
1141 CCATTTCACA AAAAAATTAT GGCAACCAAC TCAAGGTTTG TTATCTTGAA TAGTCTGGAC
1201 CCTACTAGAT CTACTGGTTT GATAATGGGT GTGCTTTTCA ATCTTTTTAC TGGTAATCTT
1261 ATGATATCTA TCGATAGTAG TATATTACAA CGACCTGGGG GCTATGAAAA TATCATTGAT
1321 AAATTTAGAG CCGACATTTT ACTGAATGAT CAGCTACAAT TGAAGCAGGT AGTTATTAAT
1381 TATTTGGAAA ATCCCGAATC AGCTTTTTCC AAGAAGCACA AAATAGATTT CAGTTGCATC
1441 AAGTCATGTT TAACCTCATG TACCACTATA GATACCGATG TAAGCGAGAT GGTTGTTCAT
1501 AAATGGCTGA AAAATTTAGG ATGTATTGAT GCACCATTTT GTTATTCTCC CATGTTAACG
1561 TTATTGGACT TTGGTGGTAT CTTTATATCT ATAAGAGATC AATTAGGAAA CCTAGAGAAC
1621 TTTCCAATAC ATAATTCAAA ACTAAGGCTT CAAAATGAAT TGTTTATAAA CCGCGAGAAA
1681 TTGAAGCTTA ATGAGGTTGA ATGTAGCATA ACTGCTATGA TTAATTCATC AAGCTCCTTC
1741 AAAGATTATC TTAAATTAGA AACATTTGGC TTCCCAATTC CTGATATCAC TTTATGTGTG
1801 GTTAATCCAG ATACGAATAC GTTGGTGCAA GACCTCACTG TTGGAGAAAT ATGGATCTCT
1861 TCTAACCACA TCACCGACGA ATTTTACCAG ATGGATAAAG TGAACGAATT TGTGTTCAAA
1921 GCAAAACTAA ATTACTCCGA AATGTTTTCT TGGGCAAAGT ATGAAATGCC TACTAACGAA
1981 AAATCGCAAG CAGTTACTGA ACAATTGGAT ACCATTCTGA ATATTTGTCC TGCAAATACC
2041 TACTTTATGA GAACCAAGCT TATGGGGTTT GTTCATAACG GAAAGATATA CGTGCTTTCA
2101 CTCATAGAAG ATATGTTTTT ACAGAATAGA TTAATCAGAT TACCTAACTG GGCTCACACA
2161 TCAAACCTTC TTTATGCCAA AAAAGGGAAT CAATCTGCTC AACCTAAAGG TAACACTGGA
2221 GCAGAAAGCA CTAAGGCTAT TGATATTTCG AGTTTAAGTG GCGAAACATC TTCGGGATAC
2281 AAAAGAGTTG TTGAATCCCA CTATCTACAA CAGATTACTG AAACAGTTGT GAGGACAGTA
2341 AATACAGTTT TTGAGGTTGC TGCATTTGAA TTACAACACC ATAAAGAAGA GCACTTTTTA
2401 GTTATGGTGG TAGAAAGCTC CCTAGCAAAA ACCGAAGAAG AGAGTAAAAA TGGCGAAACA
2461 ACAGATACTA CCTTAATGAA ATTTGCAGAA ACACAGAGGA ACAAATTAGA AACGAAAATG
2521 AATGATTTAA CTGACCAAAT TTTTAGAATT CTTTGGATTT TTCATAAAAT CCAACCAATG
2581 TGCATTTTAG TTGTGCCGAG AGATACTTTA CCTAGGAGAT ATTGTTCTCT GGAATTGGCC
2641 AATAGCACGG TAGAGAAGAA GTTTTTAAAC AATGATCTCA GTGCACAGTT TGTAAAGTTT
2701 CAATTCGATA ATGTCATATT GGATTTTTTG CCCCATTCAG CGTACTATAA TGAGAGTATA
2761 TTATCCGAGC ATTTATCAAA ATTGAGAAAG ATGGCTTTAC AAGAAGAATA CGCTATGATC
2821 GAACCAGCAT ATCGTAATGG CGGTCCCGTT AAACCAAAAC TTGCCCTACA ATGCAGTGGT
2881 GTTGATTACA GAGATGAGTC TGTTGACACT CGAAGCCATA CAAAACTCAC AGACTTCAAA
2941 TCGATTTTAG AGATTCTGGA ATGGAGAATT TCTAATTATG GGAACGAAAC GGCGTTCAGT
3001 GATGGTACAA ACACAAACCT AGTCAACTCT TCAGCTAGTA ACGATAATAA CGTTCACAAA
3061 AAGGTATCAT GGGCAAGTTT TGGTAAAATT GTCGCAGGCT TTTTGAAAAA AATCGTAGGT
3121 TCTAAAATTC CATTGAAGCA TGGTGACCCT ATTATTATTA TGTGTGAAAA TTCGGTTGAA
3181 TACGTAGCGA TGATCATGGC CTGCCTGTAC TGTAACTTAT TGGTAATTCC CCTACCAAGC
3241 GTTAAGGAGT CTGTCATAGA AGAGGACCTA AAAGGCTTGG TTAATATTAT TCAAAGTTAC
3301 AAAGTGAAAA GAGTATTTGT TGATGCTAAA TTGCACTCAT TGTTGAATGA TAATAATGTC
3361 GTAAACAAAT GTTTTAAGAA ATACAAAAGT TTGATACCCA AGATTACAGT TTTCTCAAAA
3421 GTCAAGACAA AGAATGCGTT AACAGTATCT ATGTTTAAGA ATGTGTTAAA GCAGAAATTT
3481 GGGGCCAAAC CAGGTACTAG GATTGGTATG ACACCGTGTG TTGTATGGGT AAATACAGAG
3541 TATGATGTAA CATCTAATAT TCACGTAACA ATGACACATT CGTCTTTACT GAATGCAAGC
3601 AAAATAGTCA AAGAAACTTT GCAGTTAAGA AATAATAGTC CACTTTTCTC TATATGTTCT
3661 CATACATCTG GATTGGGCTT TATGTTCAGC TGTTTGTTAG GAATTTATAC GGGTGCTTCA
3721 ACTTGCTTAT TCAGCCTTAC TGATGTTCTT ACTGACCCTA AGGAGTTTTT AATTGGCCTT
3781 CAAAATTTAA ACGTAAAAGA TTTATATTTG AAGCTTGAAA CGTTATATGC TTTACTGGAT
3841 AGAGCCTCTA GTTTGATTGA GGGATTCAAA AATAAGAAGG AGAATATAAA CTCTGCCAAA
3901 AACAATACAT CTGGCTCGCT TAGAGAAGAT GTTTTCAAGG GTGTTCGAAA TATCATGATA
3961 CCTTTTCCTA ATAGACCAAG GATTTATACA ATTGAAAATA TTTTAAAACG GTACTCAACC
4021 ATATCCCTAT CATGTACACA AATAAGTTAT GTCTATCAAC ATCACTTCAA TCCGCTTATA
4081 TCATTAAGGT CGTATCTGGA TATTCCTCCG GTTGACCTAT ATTTAGACCC ATTTTCACTG
4141 AGGGAAGGTA TAATTAGAGA AGTCAACCCC AATGATGTAA GTGCTGGAAA CTATATCAAG
4201 ATTCAAGATT CTGGTGTTGT TCCTGTATGT ACTGACGTTT CAGTAGTTAA TCCAGAGACA
4261 CTCCTACCAT GTGTTGATGG AGAATTCGGT GAGATTTGGT GCTGTTCGGA AGCTAATGCA
4321 TTCGACTACT TTGTGTGTAA CAGTTCAAAA AACAAGTTGT ACAAAGATCC CTTTATCACA
4381 GAACAATTCA AAAGTAAGAT GAAGAGCGAA GTGAATAACA CTTTAAGCTA TTTGAGAACT
4441 GGTGATCTGG GCTTTATCAA AAACGTAAGT TGCACGAATT CACAGGGAGA GGTCGTCAAT
4501 TTGAACTTGT TGTTTGTTTT GGGAAGTATT CATGAATCCA TTGAAATTCT AGGATTGACA
4561 CATTTTGTTA GTGATTTGGA GAGAACCGTT AAAGATGTTC ACAGTGACAT TGGTAGTTGT
4621 TTGATTGCCA AAGCAGGTGG ATTATTAGTG TGCTTAATTA GGTGTAAGGA ACGACATAAT
4681 CCCATCTTGG GTAATCTAAC GACTTTAATT GTTTCAGAGT TGTTAAATAA GCATGGTGTC
4741 ATTTTAGATT TATGCACATT TGTGAGAACT AAAGGTATAA GCCCAAAAAA TTCTAGTATG
4801 ATAATGGAAG TTTGGGCGAA AAATAGAGCA TCGATAATGC AAGCCTGGTT TGATCAGAAA
4861 ATTCAAATAG AAGCACAATT TGGCATAAAC TATGGTGAAA ATATTTCCAT TTATTTATTA
4921 TCAGATTATG AAAAGGACAA TATTTAA
Sequence Acquired from SGD
What is the protein sequence corresponding to your gene?
1 MDFSIPPTLP LDLQSRLNEL IQDYKDENLT RKGYETKRKQ LLDKFEISQM RPYTPLRSPN
61 SRKSKHLHRR NTSLASSITS LPNSIDRRHS IYRVTTINST SANNTPRRRS KRYTASLQSS
121 LPGSSDENGS VKDAVYNPMI PLLPRHTGAE NTSSGDSAMT DSLPLILRGR FEHYDGQTAM
181 ISINSKGKET FITWDKLYLK AERVAHELNK SHLYKMDKIL LWYNKNDVIE FTIALLGCFI
241 SGMAAVPVSF ETYSLREILE IIKVTNSKFV LISNACHRQL DNLYSSSNHS KVKLVKNDVF
301 QQIKFVKTDD LGTYTKAKKT SPTFDIPNIS YIEFTRTPLG RLSGVVMKHN ILINQFETMT
361 KILNSRSMPH WKQKSQSIRK PFHKKIMATN SRFVILNSLD PTRSTGLIMG VLFNLFTGNL
421 MISIDSSILQ RPGGYENIID KFRADILLND QLQLKQVVIN YLENPESAFS KKHKIDFSCI
481 KSCLTSCTTI DTDVSEMVVH KWLKNLGCID APFCYSPMLT LLDFGGIFIS IRDQLGNLEN
541 FPIHNSKLRL QNELFINREK LKLNEVECSI TAMINSSSSF KDYLKLETFG FPIPDITLCV
601 VNPDTNTLVQ DLTVGEIWIS SNHITDEFYQ MDKVNEFVFK AKLNYSEMFS WAKYEMPTNE
661 KSQAVTEQLD TILNICPANT YFMRTKLMGF VHNGKIYVLS LIEDMFLQNR LIRLPNWAHT
721 SNLLYAKKGN QSAQPKGNTG AESTKAIDIS SLSGETSSGY KRVVESHYLQ QITETVVRTV
781 NTVFEVAAFE LQHHKEEHFL VMVVESSLAK TEEESKNGET TDTTLMKFAE TQRNKLETKM
841 NDLTDQIFRI LWIFHKIQPM CILVVPRDTL PRRYCSLELA NSTVEKKFLN NDLSAQFVKF
901 QFDNVILDFL PHSAYYNESI LSEHLSKLRK MALQEEYAMI EPAYRNGGPV KPKLALQCSG
961 VDYRDESVDT RSHTKLTDFK SILEILEWRI SNYGNETAFS DGTNTNLVNS SASNDNNVHK
1021 KVSWASFGKI VAGFLKKIVG SKIPLKHGDP IIIMCENSVE YVAMIMACLY CNLLVIPLPS
1081 VKESVIEEDL KGLVNIIQSY KVKRVFVDAK LHSLLNDNNV VNKCFKKYKS LIPKITVFSK
1141 VKTKNALTVS MFKNVLKQKF GAKPGTRIGM TPCVVWVNTE YDVTSNIHVT MTHSSLLNAS
1201 KIVKETLQLR NNSPLFSICS HTSGLGFMFS CLLGIYTGAS TCLFSLTDVL TDPKEFLIGL
1261 QNLNVKDLYL KLETLYALLD RASSLIEGFK NKKENINSAK NNTSGSLRED VFKGVRNIMI
1321 PFPNRPRIYT IENILKRYST ISLSCTQISY VYQHHFNPLI SLRSYLDIPP VDLYLDPFSL
1381 REGIIREVNP NDVSAGNYIK IQDSGVVPVC TDVSVVNPET LLPCVDGEFG EIWCCSEANA
1441 FDYFVCNSSK NKLYKDPFIT EQFKSKMKSE VNNTLSYLRT GDLGFIKNVS CTNSQGEVVN
1501 LNLLFVLGSI HESIEILGLT HFVSDLERTV KDVHSDIGSC LIAKAGGLLV CLIRCKERHN
1561 PILGNLTTLI VSELLNKHGV ILDLCTFVRT KGISPKNSSM IMEVWAKNRA SIMQAWFDQK
1621 IQIEAQFGIN YGENISIYLL SDYEKDNI*
Sequence Acquired from SGD
What is the function of your gene?
- CMR2 functions include: Extending the lifespan of yeast bacteria, increase chemical resistance, and increase catalytic activity. Question answered with SGD
What was different about the information provided about your gene in each of the parent databases?
- Each database consists of similar information relating to CMR2. In regards to content, each database has similar information about the functions of the gene and the molecular aspects of it, but the gene identifiers are different. Each database has their own unique identifier. In addition, Ensembl also references Uniprot with the identifier name and provides a link to more information on the Uniprot page. Each database also has different research articles that tested the gene. The literature had similar concepts, but the experiments were performed by completely different scientists that may have referenced each other when completing it. In regards to the presentation of the database, NCBI and Ensembl were more complex to navigate in comparison to SGD and Uniprot. NCBI and Ensembl had more of the molecular level type information including interactions that the gene had with other genes, and had more navigation links that connected the interactions with the genes. SGD and Uniprot provided more information on the performance of the gene in a yeast cell as well as the phenotype data from it including its resistance to chemicals. The information in SGD and Uniprot were more organized and had different categories and subsections that made it easier to navigate. Question answered with SGD, Ensembl, Uniprot, and NCBI
Why did you choose your particular gene? i.e., why is it interesting to you and your partner?
- We chose the CMR2 gene because we were interested in learning about a gene that had a longer lifespan than others. In a study done on the CMR2 gene in Saccharomyces cerevisiae, Postma and other scientists found out that cells containing this gene along with a few other genes, were able to survive for about 5 years on YPD plates due to their ability to undergo long periods of low metabolic rates (Postma et al., 2009). From this paper, we were curious to research the different ways that scientists would stress this gene enough to cause an apoptotic cell death. The literature has been consistent with their research indicating that the presence of this gene in Saccharomyces cerevisiae will decrease cell death. Question answered with SGD
Acknowledgements
- We would like to thank Dr. Kam Dahlquist for the introduction to the databases utilized during this research assignment.
- Thank you to my homework partner Delisa Madere for her partnership in this assignment. We met in person and spoke over the phone in order to discuss the sharing of responsibility, contents of the page, and understanding of the assignment. All work was shared between the two of us.
- Except for what is noted above, this journal entry was done by me and not copied from another source. Jnimmers (talk) 17:29, 18 September 2019 (PDT)
- I worked with my homework partner Joey Nimmers on this assignment this week both in class and through cellular communication. We split the work evenly between us and contributed to all aspects of the project.
- We consulted the Wiki Help page when designing the format of the assignment as well. [Help Page]
- "Except for what is noted above, this individual journal entry was completed by me and not copied from another source." Dmadere (talk) 22:25, 18 September 2019 (PDT)
References
- The Ensembl Project. (n.d.). Retrieved from https://uswest.ensembl.org/Saccharomyces_cerevisiae/Info/Index
- UniProt Consortium European Bioinformatics Institute Protein Information Resource SIB Swiss Institute of Bioinformatics. (n.d.). UniProt Consortium. Retrieved from https://www.uniprot.org/
- Saccharomyces Genome Database | SGD. (n.d.). Retrieved from https://www.yeastgenome.org/
- Home - Gene - NCBI. (n.d.). Retrieved from https://www.ncbi.nlm.nih.gov/gene
- Postma, L., Lehrach, H., & Ralser, M. (2009). Surviving in the cold: yeast mutants with extended hibernating lifespan are oxidant sensitive. Aging, 1(11), 957–960. doi:10.18632/aging.100104
- Teng, X., Cheng, W. C., Qi, B., Yu, T. X., Ramachandran, K., Boersma, M. D., … Hardwick, J. M. (2011). Gene-dependent cell death in yeast. Cell death & disease, 2(8), e188. doi:10.1038/cddis.2011.72
- Wiki page week 3 assignment updates as listed by LMU BioDB 2019. (2019). Week 1. Retrieved September 18, 2019, from https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/Week_1
Biological Databases
Jnimmers
Assignment Table