Difference between revisions of "CancerSEA Week 5"
Jump to navigation
Jump to search
(→Summary Judgement: uploaded CancerSEA presentation to Wiki page) |
(→CancerSEA Presentation: linked updated presentation, fixed links) |
||
| Line 73: | Line 73: | ||
==CancerSEA Presentation== | ==CancerSEA Presentation== | ||
| − | [[ | + | [[Media:CancerSEA_presentation.pptx|CancerSEA Presentation]] |
| + | |||
| + | [[Media:Updated_CancerSEA_presentation.pptx|Updated CancerSEA Presentation]] | ||
==Acknowledgments== | ==Acknowledgments== | ||
Latest revision as of 23:15, 2 October 2019
Contents
General Info
- What is the name of the database?
- What type (or types) of database is it?
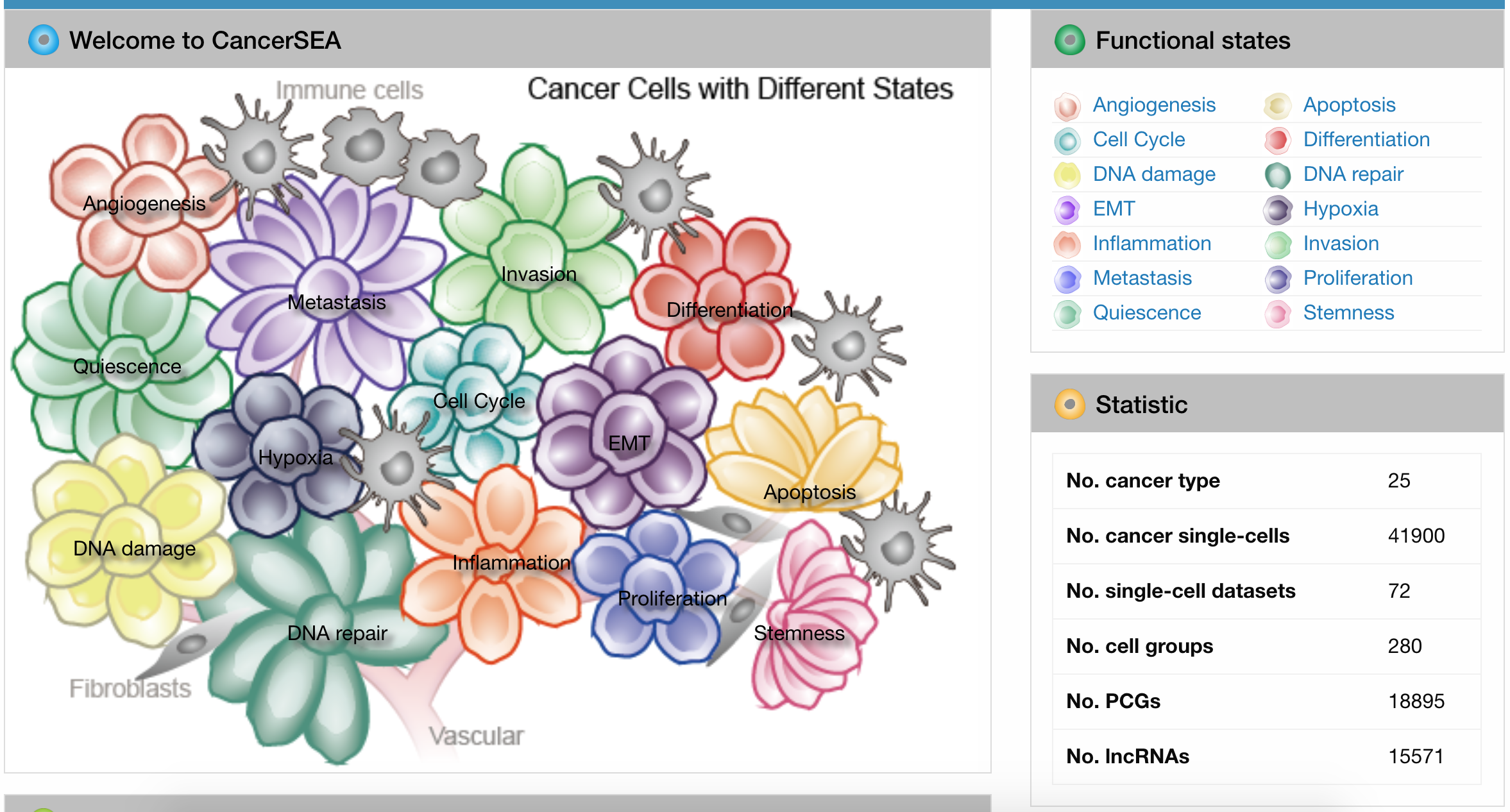
- CancerSEA is a database that decodes the various functions of cancer cells.
- What biological information (type of data) does it contain? (sequence, structure, model organism, or specialty [what?])
- -It includes the breakdown of different cancer cells that have specific functions (ie. apoptosis, DNA damage, proliferation).
- -It provides the location of where these cells are usually found in its active form.
- -It states the genes that are involved with each cell's functionality.
- What type of data source does it have? (primary versus secondary ("meta")? curated versus non-curated? if curated, is it electronic versus human curation? if human curation, is it in-house staff versus community curation?)
- -CancerSEA obtains its data through secondary sources.
- -It is curated data extracted by electronic curation.
- What individual or organization maintains the database? (public versus private? large national or multinational entity or small lab group?)
- The organization that maintains the database is the College of Bioinformatics Science and Technology, a public and international university in China.
- What is their funding source(s)?
- CancerSEA obtains funding through the college and Harbin University of Medicine.
Science Quality
- Does the content appear to completely cover its content domain?
- -The content completely covers its content domain, but as mentioned in the research article of the database, further actions include expanding the range of data to have more functional states (Huating, 2019).
- How many records does the database contain?
- -The database contains about 42,000 records of single cancer cell sets.
- What claims do the database owners make about coverage in the corresponding paper?
- -The owners claim on the paper that this database will be critical to cancer research as the sequencing of the function of cancer at the single cell is new and needed.
- -The ability to determine the heterogeneity of these cancers can be crucial to preventing excessive growth and spread of the cancers.
- What species are covered in the database? (If it is a very long list, summarize.)
- -The species that are covered within the database include carcinomas, melanomas, leukemias, or cancers that are present in different body parts, ie. cervix cancer or renal cell cancer.
- Is the database content useful? I.e., what biological questions can it be used to answer?
- -Yes. Since there is no single cure-all for all cancer cells, this data is useful for determining what kind of genes could be the target of potential medical treatments.
- Is the database content timely?
- -The database content is timely because cancer research is always needed and always useful.
- Is there a need in the scientific community for such a database at this time?
- -Yes, there is a need in the scientific community for this database at this time because not only can it be used in the educational or research setting, but it could be used for used for the creation of medical treatments. For example, medical physicians or oncologists could use it to aid in diagnoses based on what the cancer cells seem to be doing.
- Is the content covered by other databases already?
- -No, this in-depth content does not seem to be covered by other databases.
- How current is the database?
- When did the database first go online?
- -The database first went online in 2018.
- How often is the database updated? When was the last update?
- -There is currently no information as to how often the database was updated, nor is there any information as to when the last update was.
General Utility
- Are there links to other databases? Which ones?
- This database links to Ensembl, NCBI, PubMed, Gene Ontology, MSigDB, and many other larger databases.
- Is it convenient to browse the data?
- -There is a higher level of difficulty required when browsing the data since it is surrounding a complex topic. In every link that is selected, there is different data that comes up, making it difficult to keep up with the different sets, but each set has a specific header that explains what is being looked at.
- Is it convenient to download the data?
- -It is very convenient to download the data as they have a "download" tab that allows you to extract data.
- In what file formats are the data provided?
- -The data is provided in a zip file.
- What type of files, indicated by the file extension (e.g., .txt, .xml., etc.)?
- -When downloading data, the file extensions are compressed in a .zip file, and when decompressed, it is a .txt file. Another file extension used is .xls.
- Are they standard or non-standard formats? (i.e., are they following an approved standard for that type of data)?
- -The data is in standard format. The data is separated by titles that clearly represent what is expressed in the columns. The data clearly represents the data that is being asked for (i.e., Expression Profiles and Functional State Signature of Interest).
- Evaluate the “user-friendliness” of the database: can a naive user quickly navigate the website and gather useful information?
- -It might be a little difficult for someone who does not understand the content to navigate the site. There are many acronyms as well as terminologies that for the most part someone who was researching cancer cells would know. The message gets lost when navigating due to the higher level of difficulty needed when looking at this site.
- Is the website well-organized?
- -The website is easy to navigate using the links at the top of the page. On the home page, there is a diagram that allows you to pick the specific functional states of cancer cells and look them up from there. There is also an introduction to what the database is about. The database is well-organized in the way that it explains each function before clicking on it, but the information that is researched can get lost in the many links that it has. All the data is also meshed together and can get confusing when looking at it.
- Does it have a help section or tutorial?
- -There is a help section that assists users with navigation and database usage that provides screenshots of how to perform each function as well as information on what each section means.
- Are the search options sensible?
- -The search options are sensible in that it allows you to search a variety of different ways such as from inputting a gene name or list of interest, selecting the cancer type and functional state, or even selecting from the example gene list.
- Run a sample query. Do the results make sense?
- -After running a sample query on the SOX4 gene, the information that came up made sense for what was being asked. The results that came up included a brief description of the gene, the expression, and its relevance in different functional states of cancer cells, which is the purpose of the database.
- Access: Is there a license agreement or any restrictions on access to the database?
- -There is no license agreement nor is there any restrictions on access to this database. All the information is compiled on the database and has links to external pages as well for more information.
Summary Judgement
- Would you direct a colleague unfamiliar with the field to use it?
- No. This database is strictly informational for those who are specifically studying the functional states of cancer cells. There needs to be prior knowledge of the subject area. The database is very complex to navigate and understand due to the many acronyms and difficult terminologies. If the colleague had a background in the subject area, the acronyms and terms would be understood easier.
- Is this a professional or "hobby" database? The "hobby" analogy means that it was that person's hobby to make the database. It could mean that it is limited in scope, done by one or a few persons, and seems amateur.
- This database is a professional database created by a group of scientists aiming to decode the different functional states of cancer cells and genes that are relative to them. The database is not updated regularly as other professional databases are, but it does provide useful information to assist any colleague studying the functional states of cancer cells. The creators of this database are aiming to update it on a weekly basis, combine some of the single-cell data, condensing the data from the functional states, and adding new functional states in hopes to finding more diagnoses and treatments of cancer.
CancerSEA Presentation
Updated CancerSEA Presentation
Acknowledgments
- DeLisa Madere and Ivy Macaraeg set up this individual journal page on CancerSEA. We met in class to decide on the specific database and met outside of class to discuss specific details as well as to work on the presentation.
- Dr. Dhalquist for presentation instructions and rubric, as well as resources for interpreting our database.
- "Except for what is noted above, this individual journal entry was completed by me and not copied from another source." Dmadere (talk) 21:04, 30 September 2019 (PDT)
- "Except for what is noted above, this individual journal entry was completed by me and not copied from another source." Imacarae (talk) 22:06, 30 September 2019 (PDT)
References
- For access to CancerSEA database:
- - CancerSEA. (2018). College of Bioinformatics Science and Technology, Harbin Medical University. Retrieved September 30, 2019 from http://biocc.hrbmu.edu.cn/CancerSEA/home.jsp.
- - Huating Yuan et, al. CancerSEA: a cancer single-cell state atlas. (2019). Nucleic Acids Research, Volume 47, Issue D1. Retrieved on September 30, 2019 from https://academic.oup.com/nar/article/47/D1/D900/5133662
- For instructions on Week 5 assignment: LMU BioDB 2019. (2019). Week 5. Retrieved September 30, 2019, from https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/Week_5