RAD53 / YPL153C Week 3
Contents
- 1 My Favorite Gene
- 1.1 Summary of RAD53/ YPL153C:
- 1.2 1. Standard name, systematic name, and name description for your gene (from SGD)?
- 1.3 2. Gene ID (identifier) for your gene in all four databases (SGD, NCBI Gene, Ensembl, UniProt)?
- 1.4 3. DNA sequence of your gene?
- 1.5 4. Protein sequence corresponding to your gene?
- 1.6 5. Function of your gene?
- 1.7 6. What was different about the information provided about your gene in each of the parent databases?
- 1.8 Were there differences in content, the information or data itself?
- 1.9 Were there differences in presentation of the information?
- 1.10 7. Why did you choose your particular gene? i.e., why is it interesting to you and your partner?
- 1.11 8. Include an image related to your gene
- 1.12 Acknowledgments
- 1.13 References
My Favorite Gene
Christina Dominguez and Naomi Tesfaiohannes
Summary of RAD53/ YPL153C:
RAD53 is a DNA damage response. It is required for cell cycle arrest. Its signal transduction pathway component is required for DNA damage and replication. More specifically, it controls S, G1, and G2 DNA checkpoints. It is a homologue to the CHEK2 which is found in humans and can be used to study breast cancer.
1. Standard name, systematic name, and name description for your gene (from SGD)?
Standard Name: Rad53
Systematic Name: YPL153C
Name Description: RADiation sensitive
It is a DNA damage response kinase and plays a role in the initiation of DNA replication.
2. Gene ID (identifier) for your gene in all four databases (SGD, NCBI Gene, Ensembl, UniProt)?
Gene ID in SGD: S000006074
Gene ID in NCBI: S000006074
Gene ID in Ensembl: S000006074
Gene ID in UniProt: S000006074
All 4 databases use the same Gene ID
3. DNA sequence of your gene?
ATGGAAAATATTACACAACCCACACAGCAATCCACGCAGGCTACTCAAAGGTTTTTGATT GAGAAGTTTTCTCAAGAACAGATCGGCGAAAACATTGTGTGCAGGGTCATTTGTACCACG GGTCAAATTCCCATCCGAGATTTGTCAGCTGATATTTCACAAGTGCTTAAGGAAAAACGA TCCATAAAGAAAGTTTGGACATTTGGTAGAAACCCAGCCTGTGACTATCATTTAGGAAAC ATTTCAAGACTGTCAAATAAGCATTTCCAAATACTACTAGGAGAAGACGGTAACCTTTTA TTGAATGACATTTCCACTAATGGGACCTGGTTAAATGGGCAAAAAGTCGAGAAGAACAGC AATCAGTTACTGTCTCAAGGTGATGAAATAACCGTTGGTGTAGGCGTGGAATCAGATATT TTATCTCTGGTCATTTTCATAAACGACAAATTTAAGCAGTGCCTCGAGCAGAACAAAGTT GATCGCATAAGATCTAACCTGAAAAATACCTCTAAAATAGCTTCTCCTGGTCTTACATCA TCTACTGCATCATCAATGGTGGCCAACAAGACTGGTATTTTTAAGGATTTTTCGATTATT GACGAAGTGGTGGGCCAGGGTGCATTTGCCACAGTAAAGAAAGCCATTGAAAGAACTACT GGGAAAACATTCGCGGTGAAGATTATAAGTAAACGCAAAGTAATAGGCAATATGGATGGT GTGACAAGAGAGTTAGAAGTATTGCAAAAGCTCAATCATCCAAGGATAGTACGATTGAAA GGATTTTATGAAGATACTGAGAGTTATTATATGGTGATGGAGTTCGTTTCTGGTGGTGAC TTAATGGATTTTGTTGCTGCTCATGGTGCGGTTGGAGAAGATGCTGGGAGGGAGATATCC AGGCAGATACTCACAGCAATAAAATACATTCACTCTATGGGCATCAGCCATCGTGACCTA AAGCCCGATAATATTCTTATTGAACAAGACGATCCTGTATTGGTAAAGATAACCGACTTT GGTCTGGCAAAAGTACAAGGAAATGGGTCTTTTATGAAAACCTTCTGTGGCACTTTGGCA TATGTGGCACCTGAAGTCATCAGAGGTAAAGATACATCCGTATCTCCTGATGAATACGAA GAAAGGAATGAGTACTCTTCGTTAGTGGATATGTGGTCAATGGGATGTCTTGTGTATGTT ATCCTAACGGGCCACTTACCTTTTAGTGGTAGCACACAGGACCAATTATATAAACAGATT GGAAGAGGCTCATATCATGAAGGGCCCCTCAAAGATTTCCGGATATCTGAAGAAGCAAGA GATTTCATAGATTCATTGTTACAGGTGGATCCAAATAATAGGTCGACAGCTGCAAAAGCC TTGAATCATCCCTGGATCAAGATGAGTCCATTGGGCTCACAATCATATGGTGATTTTTCA CAAATATCCTTATCACAATCGTTGTCGCAGCAGAAATTATTAGAAAATATGGACGATGCT CAATACGAATTTGTCAAAGCGCAAAGGAAATTACAAATGGAGCAACAACTTCAAGAACAG GATCAGGAAGACCAAGATGGAAAAATTCAAGGATTTAAAATACCCGCACACGCCCCTATT CGATATACACAGCCCAAAAGCATTGAAGCAGAAACTAGAGAACAAAAACTTTTACATTCC AATAATACTGAGAATGTCAAGAGCTCAAAGAAAAAGGGTAATGGTAGGTTTTTAACTTTA AAACCATTGCCTGACAGCATTATTCAAGAAAGCCTGGAGATTCAGCAAGGTGTGAATCCA TTTTTCATTGGTAGATCCGAGGATTGCAATTGTAAAATTGAAGACAATAGGTTGTCTCGA GTTCATTGCTTCATTTTCAAAAAGAGGCATGCTGTAGGCAAAAGCATGTATGAATCTCCG GCACAAGGTTTAGATGATATTTGGTATTGCCACACCGGAACTAACGTGAGCTATTTAAAT AATAACCGCATGATACAGGGTACGAAATTCCTTTTACAAGACGGAGATGAAATCAAGATC ATTTGGGATAAAAACAATAAATTTGTCATTGGCTTTAAAGTGGAAATTAACGATACTACA GGTCTGTTTAACGAGGGATTAGGTATGTTACAAGAACAAAGAGTAGTACTTAAGCAAACA GCCGAAGAAAAAGATTTGGTGAAAAAGTTAACCCAGATGATGGCAGCTCAACGTGCAAAT CAACCCTCGGCTTCTTCTTCATCAATGTCGGCTAAGAAGCCGCCAGTTAGCGATACAAAT AATAACGGCAATAATTCGGTACTAAACGACTTGGTAGAGTCACCGATTAATGCGAATACG GGGAACATTTTGAAGAGAATACATTCGGTAAGTTTATCGCAATCACAAATTGATCCTAGT AAGAAGGTTAAAAGGGCAAAATTGGACCAAACCTCAAAAGGCCCCGAGAATTTGCAATTT TCGTAA
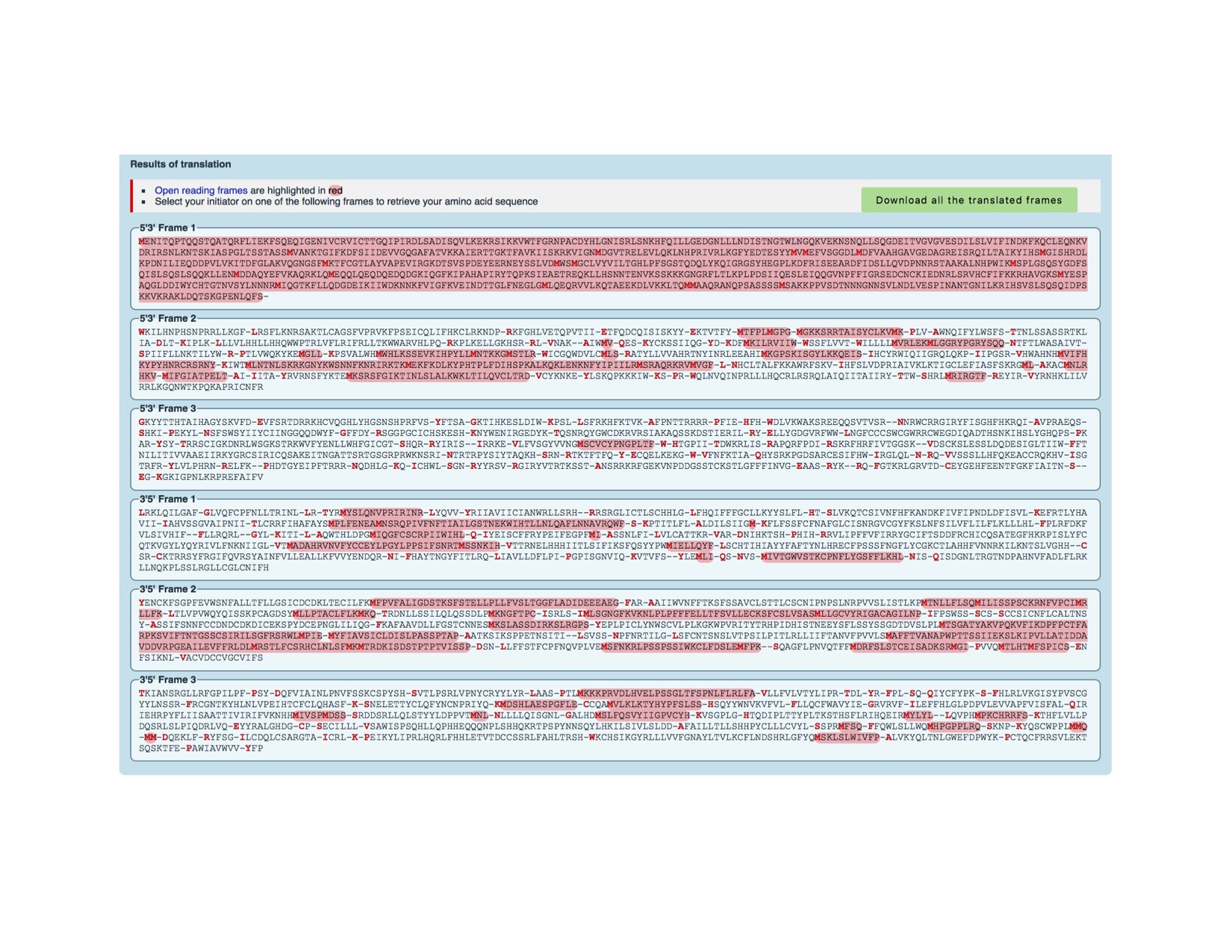
4. Protein sequence corresponding to your gene?
Frame 1 5'3' encodes the entire protein sequence
https://web.expasy.org/translate/
5. Function of your gene?
RAD53 encodes for a protein kinase which is required for the cell cycle checkpoint. It amplifies the initial signals for proteins that recognize DNA damage. A loss in RAD53 can lead to defects in checkpoint activation.
6. What was different about the information provided about your gene in each of the parent databases?
The UniProt database gave the most detail about where it functions in DNA damage checkpoints. SGD itself and Ensembl provided many more generalized functions that the genes are believed to be involved in without going into much detail. NCBI gave more information about mapping the gene and wasn't as straight forward about the functions.
Were there differences in content, the information or data itself?
The difference in content was mainly in the more generalized or more specific functions given on the databases. For example, Uniprot gave detailed information specific to the gene's involvement with DNA damage checkpoints. Although the other databases did mention its involvement with DNA damage, it did not go into specifics but gave more functions that it is involved with. All databases were consistent with the gene's use in the study of breast cancer.
Were there differences in presentation of the information?
NCBI did the best job at presenting the information so that you could scroll through one page and see all the data there was to offer. It was the most difficult to understand what the gene's role was. SGD and UnitProt had the best visual view of the gene and information. Ensembl was the most confusing to navigate in terms of different links to click on to find more information.
7. Why did you choose your particular gene? i.e., why is it interesting to you and your partner?
We wanted to research a gene that was relatable to the field of health and the pursuit of eradicating a disease, such as cancer. We chose the RAD53/YPL153C gene because of its involvement in the study of breast cancer. It being homologous to human CHEK2 has allowed it to be used in learning more about the nature of breast cancer and treatment.
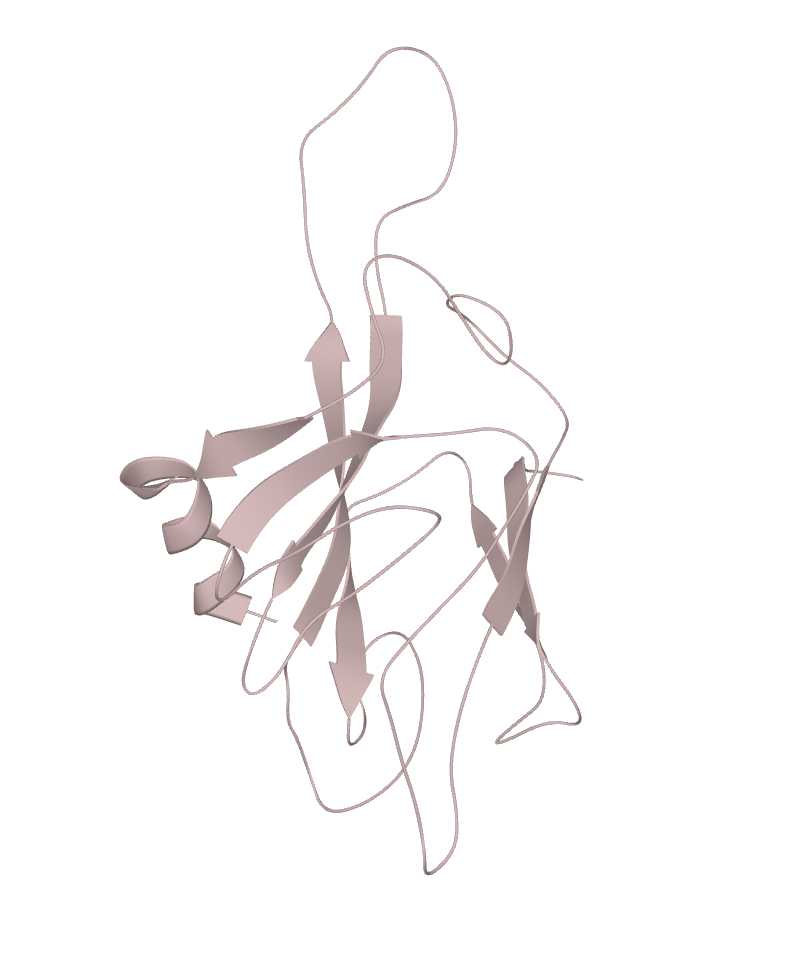
Image of the RAD53 gene
Acknowledgments
- We worked together during multiple classes and met once outside of class to collaborate on the different databases information. For the beginning of this assignment, work was saved on the wiki page Cdomin12 and Ntesfaio Week 3. However, the name of the week 3 individual assignment was to be named after the gene. The proper name was then changed to RAD53 / YPL153C Week 3 and the syntax that was edited on the previous page was transferred over to this page. Previous saves can be seen in the "view history" tab of the page Cdomin12 and Ntesfaio Week 3.
"Except for what is noted above, this individual journal entry was completed by me and not copied from another source." Ntesfaio (talk) 16:34, 16 September 2019 (PDT)
"Except for what is noted above, this individual journal entry was completed by me and not copied from another source." Cdomin12 (talk) 21:10, 17 September 2019 (PDT)
References
- ExPASy Translator. Retrieved September 17,2019 from [EXPASy]
- SGD Database. Retrieved September 17,2019 from [SGD Rad53]
- NCBI Database. Retrieved September 17,2019 from [NCBI Rad53]
- UniProt Database. Retrieved September 17, 2019 from [Rad53]
- Ensembl Database. Retrieved September 17, 2019 from [Rad53]
- Week 3 Assignment page is: LMU BioDB 2019. (2019). Week 3. Retrieved September 17, 2019 from [Week 3]
Ntesfaio Final Individual Reflection