Icrespin Journal Week 6
Electronic Lab Notebook
Purpose
The purpose of this assignment is to improve the skills on making an electronic lab notebook. In addition, articles were read so that the user can better understand how experiments on DNA microarrays function. Another important purpose is to view analysis problems regarding this and discussing research ethics.
Questions
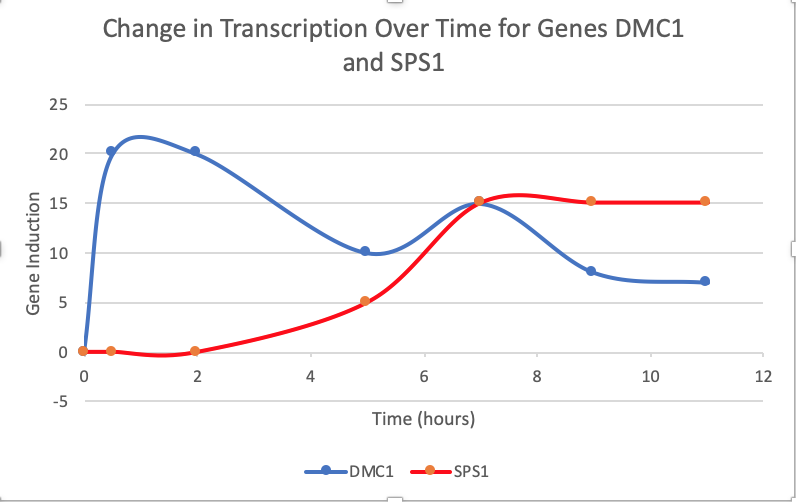
- (Question 5, p. 110) Choose two genes from Figure 4.6b (PDF of figures on Brightspace) and draw a graph to represent the change in transcription over time. You can either create your plot in Excel and put the image up on your wiki page or you can do it by hand and upload a picture or scan.
- (Question 6b, p. 110) Look at Figure 4.7, which depicts the loss of oxygen over time and the transcriptional response of three genes. These data are the ratios of transcription for genes X, Y, and Z during the depletion of oxygen. Using the color scale from Figure 4.6, determine the color for each ratio in Figure 4.7b. (Use the nomenclature "bright green", "medium green", "dim green", "black", "dim red", "medium red", or "bright red" for your answers.)
| 1 Hour | 3 Hours | 5 Hours | 9 Hours | |
|---|---|---|---|---|
| Gene X | Black | Medium Red | Black | Bright Green |
| Gene Y | Black | Bright Red | Black | Bright Green |
| Gene Z | Black | Dim Red | Medium Red | Medium Red |
- (Question 7, p. 110) Were any of the genes in Figure 4.7b transcribed similarly? If so, which ones were transcribed similarly to which ones?
- Gene X and Y were transcribed similarly. At first, all of them start out the same, but later Gene Z is transcribed very differently. Both X and Y go up with slightly different colors than go back to the same color as hour 1. By 9 hours, they are both the same.
- (Question 9, p. 118) Why would most spots be yellow at the first time point? I.e., what is the technical reason that spots show up as yellow - where does the yellow color come from? And, what would be the biological reason that the experiment resulted in most spots being yellow?
- The reason that there are yellow spots is because it's indicating that they're merged together. Using cDNA, the mircroassays indicate colors on a scale thats runs between red and green. Since some dyes are hard to separate, the system begins reading it as one whole object. This creates the yellow spots.
- (Question 10, p. 118) Go to the Saccharomyces Genome Database and search for the gene TEF4; you will see it is involved in translation. Look at the time point labeled OD 3.7 in Figure 4.12, and find the TEF4 spot. Over the course of this experiment, was TEF4 induced or repressed? Hypothesize why TEF4’s change in expression was part of the cell’s response to a reduction in available glucose (i.e., the only available food).
- TEF4 consist of a green color, which states that it is being repressed. Due to this, TEF4's change in expression was part of the cell's response to a reduction, since it plays a role in elongation. Because of this, it causes limitations with the cell's response. Due to these limitations, there isn't sufficient production of glucose, which causes the use of stored energy.
- (Question, 11, p. 120) Why would TCA cycle genes be induced if the glucose supply is running out?
- TCA genes would be induced because glucose is the basis of glycolysis. With a low supply of glucose, there wouldn't be enough to convert it pyruvate. Without this conversion, TCA cannot continue to its next stages of using ATP and NADH.
- (Question 12, p. 120) What mechanism could the genome use to ensure genes for enzymes in a common pathway are induced or repressed simultaneously?
- The mechanism that the genome could use would be the "guilt by association" method. It consist of two rules that state genes with similar expression profiles should have similar promoters and vice versa. By using this method, it would give information of how similar genes can help predict or discover the unknown ones.
- (Question 13, p. 121) Consider a microarray experiment where cells deleted for the repressor TUP1 were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented glucose-repressed genes to be in the later time points of this experiment?
- In the microarray, there would be spots consisting of a gradual change from green to red. The spots would represent those different shades of those colors showing how glucose has depleted over time.
- (Question 14, p. 121) Consider a microarray experiment where cells that overexpress the transcription factor Yap1p were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented Yap1p target genes to be in the later time points of this experiment?
- For a transcription factor that is being overexpressed, it would start in red and end in green. Over time, there would be a huge increase of glucose. From the t0 to a later time, there would be different shades heading towards green.
- (Question 16, p. 121) Using the microarray data, how could you verify that you had truly deleted TUP1 or overexpressed YAP1 in the experiments described in questions 8 and 9?
- In order to verify that it has been truly deleted, an analysis can be conducted to see changes of gene expressions. The analysis would consist of spots showing those changes. If there is a change between a regular gene expression and one that has been deleted/overexpressed, then it would verify the hypotheses.
Acknowledgments
- This week, my homework partner was Ivy Macaraeg. We exchanged phone numbers and discussed for a little while about the objectives of this week's assignment.
- References (as shown under References section) were copied from Week's 6 Assignment page.
- "Except for what is noted above, this individual journal entry was completed by me and not copied from another source."Icrespin (talk) 21:01, 9 October 2019 (PDT)
References
Alberts et al. (2002) Molecular Biology of the Cell, Ch. 8: Microarrays
Brown, P.O. & Botstein, D. (1999) Exploring the new world of the genome with DNA microarrays Nature Genetics 21: 33-37.
Campbell, A.M. and Heyer, L.J. (2003), “Chapter 4: Basic Research with DNA Microarrays”, in Discovering Genomics, Proteomics, and Bioinformatics, Cold Spring Harbor Laboratory Press, pp. 107-124. (Available on Brightspace)
DeRisi, J.L., Iyer, V.R., and Brown, P.O. (1997) Exploring the Metabolic and Genetic Control of Gene Expression on a Genomic Scale. Science 278: 680-686.
LMU BioDB 2019. (2019). Week 6. Retrieved October 9, 2019, from https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/Week_6