Difference between revisions of "Ancient mtDNA Week 5"
Jump to navigation
Jump to search
(added my template to the page) |
(added presentation) |
||
| (53 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | ==Authors== | ||
| + | {{Template:msamdars}} | ||
{{Template:eyoung20}} | {{Template:eyoung20}} | ||
| + | ==Purpose== | ||
| + | The purpose of this assignment is to critically analyze a database and be able to present our finding in an oral format. | ||
| + | ==Database Evaluation== | ||
| + | ===General information about the database=== | ||
| + | #What is the name of the database? | ||
| + | #*[https://amtdb.org/ Ancient mtDNA Database] | ||
| + | #*Database was chosen from the [https://academic.oup.com/nar/issue/47/D1 ''Nucleic Acids Research'' Database Issue Table of Contents 2019]. | ||
| + | #What type (or types) of database is it? | ||
| + | #*It is a Database of Ancient mitochondrial DNA. The Database is built up of secondary source information curated by the research team under Dr. Edvard Ehler from the Laboratory of Genomics and Bioinformatics in Institute of Molecular Genetics at the Czech Academy of Sciences, Prague, Czech Republic, working with a team from Anna Juras ancient DNA lab, Institute of Anthropology, UAM, Poznan, Poland. | ||
| + | #** Information on data sources found in the journal article [https://academic.oup.com/nar/article/47/D1/D29/5106144?searchresult=1/ AmtDB: a database of ancient human mitochondrial genomes] or on database website [https://amtdb.org/ Ancient mtDNA] by clicking the tab FAQ & Help then viewing the references section. The Information on who currated the database can be found on the database website [https://amtdb.org/ Ancient mtDNA] under the contact tab. | ||
| + | #What individual or organization maintains the database? | ||
| + | #* The Database is maintained by the research team under Dr. Edvard Ehler from the Laboratory of Genomics and Bioinformatics in Institute of Molecular Genetics at the Czech Academy of Sciences, Prague, Czech Republic, working with a team from Anna Juras ancient DNA lab, Institute of Anthropology, UAM, Poznan, Poland. | ||
| + | #** This information was found on the [https://amtdb.org/ Ancient mtDNA] database site under the contact tab. | ||
| + | #What is their funding source | ||
| + | #*"Ministry of Education, Youth and Sports of the Czech Republic [ELIXIR-CZ project LM2015047, part of the international ELIXIR infrastructure, under the Projects CESNET, LM2015042]; Polish National Science Center [2014/12/W/NZ2/00466]. Funding for open access charge: Ministry of Education, Youth and Sports of the Czech Republic" | ||
| + | #** Information found on the journal article [https://academic.oup.com/nar/article/47/D1/D29/5106144?searchresult=1/ AmtDB: a database of ancient human mitochondrial genomes] | ||
| + | |||
| + | ===Scientific quality of the database=== | ||
| + | # Does the content appear to completely cover its content domain? | ||
| + | #* The database contains 1314 records on ancient human mitochondrial DNA according to their [https://amtdb.org About page]. The database paper does not make any claim about how much of the literature regarding ancient human mitochondrial DNA that it covers (Ehler et al., 2018). However, the map that it allows users to browse data by only shows availability of samples from the Eurasian continent (primarily the Mid-Western and Russian parts). Although I do not know too much about biological archaeology, I am not sure that ancient human DNA samples exist from only those locations, and thus do not believe that the database is complete | ||
| + | # What species are covered in the database? | ||
| + | #* The database only claims to cover ancient ''Homo sapiens''. | ||
| + | # Is the database content useful? I.e., what biological questions can it be used to answer? | ||
| + | #* The database can be used to access resources regarding ancient human mitochondrial DNA. This information is useful in uncovering information regarding genetic variation among human DNA and analyzing population data regarding ancient humans. (Ramakrishnan & Hadley, 2009) | ||
| + | # Is the database content timely? | ||
| + | #* No other database for this type of data exists currently, but I am not certain how common forensic studies of ancient human DNA is. This database is in a unique position. Other databases contain ancient DNA, mitochondrial DNA, ancient mitochondrial DNA, but there are no other databases that are specific to ancient human mitochondrial DNA. | ||
| + | # How ''current'' is the database? | ||
| + | #* The database just launched in first launched September 5, 2018 (European date format?) taken from [https://amtdb.org/help#div-tab-log this] log of the database, and thus contains relatively up-to-date ancient mitochondrial human DNA samples. Since its launch according to the log, the database has been updated three times with the last update being on May 9th of 2019. | ||
| + | |||
| + | ===General utility of the database to the scientific community=== | ||
| + | #Are there links to other databases? Which ones? | ||
| + | #*The site is linked to [https://www.elixir-czech.cz/ ELIXER Czech Republic] | ||
| + | #*It is also linked to the [https://www.img.cas.cz/en/ Institute of Molecular Genetics of the Czech Academy of Sciences] website | ||
| + | #Is it convenient to browse the data? | ||
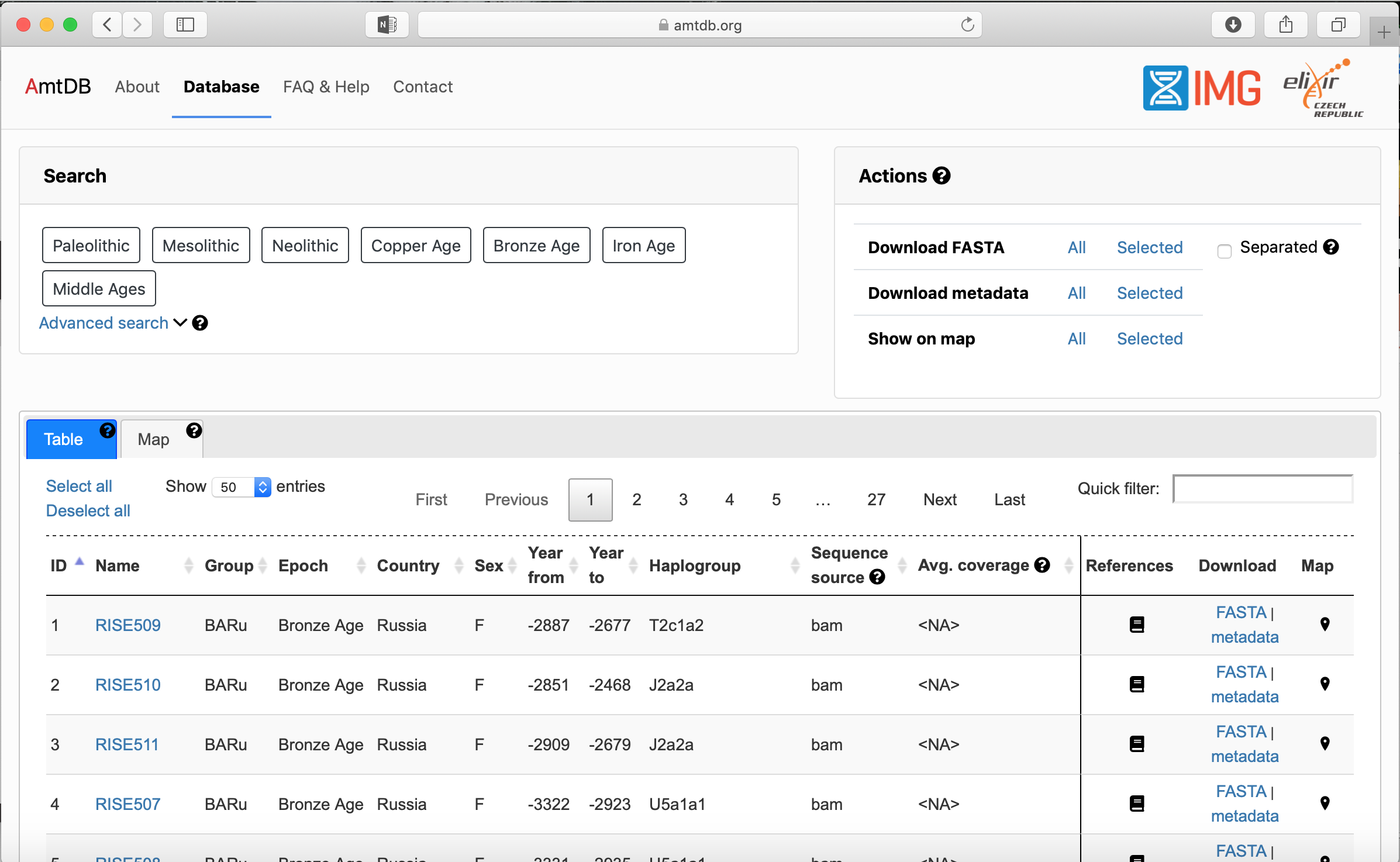
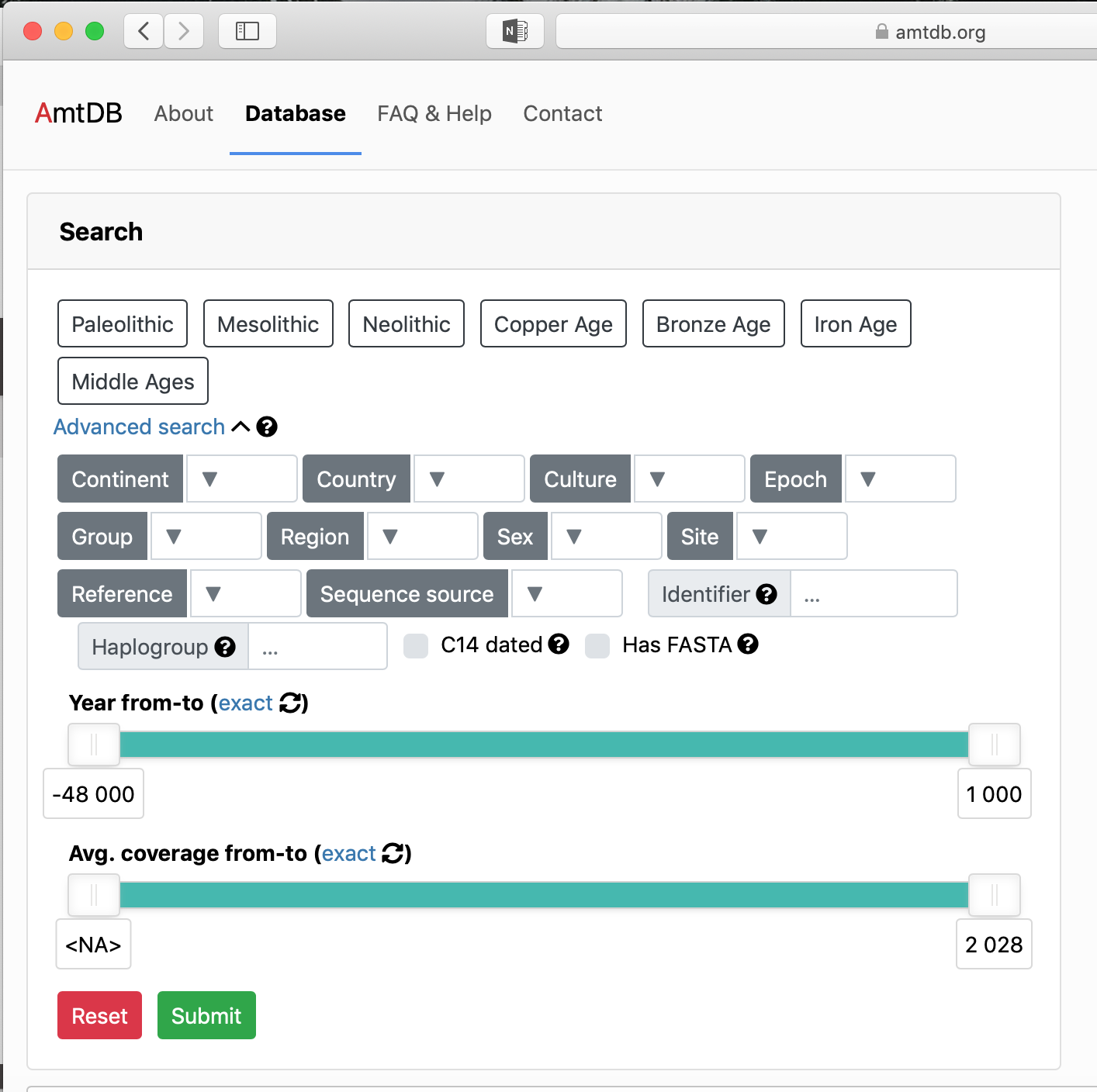
| + | #*It is very easy to browse data on this site. The data is able to be searched, advanced searched, and view by location on a map. [[File:AmtDB_Databasepage.png|thumb|200px|Search Options]] [[File:AmtDB_Advancedsearch.png|thumb|200px|Advanced Search Options ]] | ||
| + | #Is it convenient to download data? | ||
| + | #* Downloading data is fairly easy and there is a clear way under the actions option to do just that. You can even select which sequences you would like and then only download those. | ||
| + | #*The Data can be downloaded under standard formats. DNA sequences may be downloaded as FASTA sequences (.fasta) for DNA sequences or for other data in a tabular format as a comma-separated values (.csv) file. | ||
| + | #Evaluate the “user-friendliness” of the database: can a naive user quickly navigate the website and gather useful information? | ||
| + | #* The web site is well organized and very easy to navigate. This is probably do to the fact that the web design is clean and simple. | ||
| + | #*There is FAQ & Help tab on the page that has FAQs, Metadata information, references, a log of edits to the site, and the terms of service. However there is no help with how the information can be used once downloaded or what systems it is compatible with. | ||
| + | #*There are many search options on the site itself that allow you to easily narrow down the parameters, these search options also work on the map as well, so there can be a visual relationship to the data. | ||
| + | #*The run of the sample query was somewhat pointless, the data is exported in one large table. The query is however useful in narrowing down the information you want to look at. The same task can be done on the advanced search on the site itself. However this would be very useful if you wanted to compare the mitochondrial DNA form another Database with this AmtDB. | ||
| + | #Access: Is there a license agreement or any restrictions on access to the database? | ||
| + | #* there are no agreements or restrictions on the access to this database, just a disclaimer under terms of service, which is located under the FAQ & Help tab. | ||
| + | |||
| + | ===Summary judgment=== | ||
| + | # Would you direct a colleague unfamiliar with the field to use it? | ||
| + | #* I would not direct a colleague unfamiliar with the biology field to use the database. However, the database is very usable to anyone lightly familiar with humans and databases with DNA sequences. The database provides the ability to do meta-analysis via the implementation of an option to download all data. Furthermore, it is extremely informative and provides information that is intriguing, including the ability to sort data by estimated dates of living. The oldest data entry appears to be from nearly 48,000 years ago. | ||
| + | #Is this a professional or "hobby" database? The "hobby" analogy means that it was that person's hobby to make the database. It could mean that it is limited in scope, done by one or a few persons, and seems amateur. | ||
| + | #* This seems to be a fully-funded, professional database that is fully capable of scaling up and adapting to its users' needs. It seems to have been updated fairly recently, and thus it seems that its creators and distributors are truly invested in its maintenance. | ||
| + | |||
| + | ==Presentation== | ||
| + | [[File:Ancient_mtDNA_Presentation_.pdf]] | ||
| + | |||
| + | ==Acknowledgements== | ||
| + | This week, the two partners for this project are [[User:msamdars|Mihir Samdarshi]] and [[User:eyoung20|Emma Young]]. Both partners worked with each other outside of class to assist each other in finding and synthesizing the necessary information required to complete this assignment. | ||
| + | |||
| + | We would like to acknowledge [[user:Kdahlquist]] for her instruction on this topic and how to give a presentation. | ||
| + | |||
| + | Except for what is noted above, this individual journal entry was completed by us and not copied from another source. | ||
| + | |||
| + | [[User:Eyoung20|Eyoung20]] ([[User talk:Eyoung20|talk]]) 20:54, 30 September 2019 (PDT) | ||
| + | [[User:Msamdars|Msamdars]] ([[User talk:Msamdars|talk]]) 23:35, 30 September 2019 (PDT) | ||
| + | |||
| + | ==References== | ||
| + | # Ehler, E., Novotný, J., Juras, A., Chyleński, M., Moravčík, O., & Pačes, J. (2018). AmtDB: a database of ancient human mitochondrial genomes. Nucleic acids research, 47(D1), D29-D32. | ||
| + | # Ramakrishnan, U., & Hadly, E. A. (2009). Using phylochronology to reveal cryptic population histories: review and synthesis of 29 ancient DNA studies. Molecular Ecology, 18(7), 1310-1330. | ||
| + | # LMU BioDB 2019. (2019). Week 5. Retrieved September 30, 2019, from https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/Week_5 | ||
| + | # MediaWiki (2019). Category: Help. Retrieved September 30, 2019, from https://www.mediawiki.org/wiki/Category:Help | ||
Latest revision as of 22:59, 30 September 2019
Contents
Authors
| Links | ||
|---|---|---|
| Mihir Samdarshi User Page | ||
| Assignment Pages | Personal Journal Entries | Shared Journal Entries |
| Week 1 | Journal Week 1 | Class Journal Week 1 |
| Week 2 | Journal Week 2 | Class Journal Week 2 |
| Week 3 | FAS2/YPL231W Week 3 | Class Journal Week 3 |
| Week 4 | Journal Week 4 | Class Journal Week 4 |
| Week 5 | Database - AmtDB | Class Journal Week 5 |
| Week 6 | Journal Week 6 | Class Journal Week 6 |
| Week 7 | Journal Week 7 | Class Journal Week 7 |
| Week 8 | Journal Week 8 | Class Journal Week 8 |
| Week 9 | Journal Week 9 | Class Journal Week 9 |
| Assignment Pages | Personal Journal Entries | |
| Week 10 | Journal Week 10 | |
| Week 11 | Journal Week 11 | |
| Week 12/13 | Journal Week 12/13 | |
| Team Project Links | ||
| Skinny Genes Team Page | ||
Purpose
The purpose of this assignment is to critically analyze a database and be able to present our finding in an oral format.
Database Evaluation
General information about the database
- What is the name of the database?
- Ancient mtDNA Database
- Database was chosen from the Nucleic Acids Research Database Issue Table of Contents 2019.
- What type (or types) of database is it?
- It is a Database of Ancient mitochondrial DNA. The Database is built up of secondary source information curated by the research team under Dr. Edvard Ehler from the Laboratory of Genomics and Bioinformatics in Institute of Molecular Genetics at the Czech Academy of Sciences, Prague, Czech Republic, working with a team from Anna Juras ancient DNA lab, Institute of Anthropology, UAM, Poznan, Poland.
- Information on data sources found in the journal article AmtDB: a database of ancient human mitochondrial genomes or on database website Ancient mtDNA by clicking the tab FAQ & Help then viewing the references section. The Information on who currated the database can be found on the database website Ancient mtDNA under the contact tab.
- It is a Database of Ancient mitochondrial DNA. The Database is built up of secondary source information curated by the research team under Dr. Edvard Ehler from the Laboratory of Genomics and Bioinformatics in Institute of Molecular Genetics at the Czech Academy of Sciences, Prague, Czech Republic, working with a team from Anna Juras ancient DNA lab, Institute of Anthropology, UAM, Poznan, Poland.
- What individual or organization maintains the database?
- The Database is maintained by the research team under Dr. Edvard Ehler from the Laboratory of Genomics and Bioinformatics in Institute of Molecular Genetics at the Czech Academy of Sciences, Prague, Czech Republic, working with a team from Anna Juras ancient DNA lab, Institute of Anthropology, UAM, Poznan, Poland.
- This information was found on the Ancient mtDNA database site under the contact tab.
- The Database is maintained by the research team under Dr. Edvard Ehler from the Laboratory of Genomics and Bioinformatics in Institute of Molecular Genetics at the Czech Academy of Sciences, Prague, Czech Republic, working with a team from Anna Juras ancient DNA lab, Institute of Anthropology, UAM, Poznan, Poland.
- What is their funding source
- "Ministry of Education, Youth and Sports of the Czech Republic [ELIXIR-CZ project LM2015047, part of the international ELIXIR infrastructure, under the Projects CESNET, LM2015042]; Polish National Science Center [2014/12/W/NZ2/00466]. Funding for open access charge: Ministry of Education, Youth and Sports of the Czech Republic"
- Information found on the journal article AmtDB: a database of ancient human mitochondrial genomes
- "Ministry of Education, Youth and Sports of the Czech Republic [ELIXIR-CZ project LM2015047, part of the international ELIXIR infrastructure, under the Projects CESNET, LM2015042]; Polish National Science Center [2014/12/W/NZ2/00466]. Funding for open access charge: Ministry of Education, Youth and Sports of the Czech Republic"
Scientific quality of the database
- Does the content appear to completely cover its content domain?
- The database contains 1314 records on ancient human mitochondrial DNA according to their About page. The database paper does not make any claim about how much of the literature regarding ancient human mitochondrial DNA that it covers (Ehler et al., 2018). However, the map that it allows users to browse data by only shows availability of samples from the Eurasian continent (primarily the Mid-Western and Russian parts). Although I do not know too much about biological archaeology, I am not sure that ancient human DNA samples exist from only those locations, and thus do not believe that the database is complete
- What species are covered in the database?
- The database only claims to cover ancient Homo sapiens.
- Is the database content useful? I.e., what biological questions can it be used to answer?
- The database can be used to access resources regarding ancient human mitochondrial DNA. This information is useful in uncovering information regarding genetic variation among human DNA and analyzing population data regarding ancient humans. (Ramakrishnan & Hadley, 2009)
- Is the database content timely?
- No other database for this type of data exists currently, but I am not certain how common forensic studies of ancient human DNA is. This database is in a unique position. Other databases contain ancient DNA, mitochondrial DNA, ancient mitochondrial DNA, but there are no other databases that are specific to ancient human mitochondrial DNA.
- How current is the database?
- The database just launched in first launched September 5, 2018 (European date format?) taken from this log of the database, and thus contains relatively up-to-date ancient mitochondrial human DNA samples. Since its launch according to the log, the database has been updated three times with the last update being on May 9th of 2019.
General utility of the database to the scientific community
- Are there links to other databases? Which ones?
- The site is linked to ELIXER Czech Republic
- It is also linked to the Institute of Molecular Genetics of the Czech Academy of Sciences website
- Is it convenient to browse the data?
- It is very easy to browse data on this site. The data is able to be searched, advanced searched, and view by location on a map.
- Is it convenient to download data?
- Downloading data is fairly easy and there is a clear way under the actions option to do just that. You can even select which sequences you would like and then only download those.
- The Data can be downloaded under standard formats. DNA sequences may be downloaded as FASTA sequences (.fasta) for DNA sequences or for other data in a tabular format as a comma-separated values (.csv) file.
- Evaluate the “user-friendliness” of the database: can a naive user quickly navigate the website and gather useful information?
- The web site is well organized and very easy to navigate. This is probably do to the fact that the web design is clean and simple.
- There is FAQ & Help tab on the page that has FAQs, Metadata information, references, a log of edits to the site, and the terms of service. However there is no help with how the information can be used once downloaded or what systems it is compatible with.
- There are many search options on the site itself that allow you to easily narrow down the parameters, these search options also work on the map as well, so there can be a visual relationship to the data.
- The run of the sample query was somewhat pointless, the data is exported in one large table. The query is however useful in narrowing down the information you want to look at. The same task can be done on the advanced search on the site itself. However this would be very useful if you wanted to compare the mitochondrial DNA form another Database with this AmtDB.
- Access: Is there a license agreement or any restrictions on access to the database?
- there are no agreements or restrictions on the access to this database, just a disclaimer under terms of service, which is located under the FAQ & Help tab.
Summary judgment
- Would you direct a colleague unfamiliar with the field to use it?
- I would not direct a colleague unfamiliar with the biology field to use the database. However, the database is very usable to anyone lightly familiar with humans and databases with DNA sequences. The database provides the ability to do meta-analysis via the implementation of an option to download all data. Furthermore, it is extremely informative and provides information that is intriguing, including the ability to sort data by estimated dates of living. The oldest data entry appears to be from nearly 48,000 years ago.
- Is this a professional or "hobby" database? The "hobby" analogy means that it was that person's hobby to make the database. It could mean that it is limited in scope, done by one or a few persons, and seems amateur.
- This seems to be a fully-funded, professional database that is fully capable of scaling up and adapting to its users' needs. It seems to have been updated fairly recently, and thus it seems that its creators and distributors are truly invested in its maintenance.
Presentation
File:Ancient mtDNA Presentation .pdf
Acknowledgements
This week, the two partners for this project are Mihir Samdarshi and Emma Young. Both partners worked with each other outside of class to assist each other in finding and synthesizing the necessary information required to complete this assignment.
We would like to acknowledge user:Kdahlquist for her instruction on this topic and how to give a presentation.
Except for what is noted above, this individual journal entry was completed by us and not copied from another source.
Eyoung20 (talk) 20:54, 30 September 2019 (PDT) Msamdars (talk) 23:35, 30 September 2019 (PDT)
References
- Ehler, E., Novotný, J., Juras, A., Chyleński, M., Moravčík, O., & Pačes, J. (2018). AmtDB: a database of ancient human mitochondrial genomes. Nucleic acids research, 47(D1), D29-D32.
- Ramakrishnan, U., & Hadly, E. A. (2009). Using phylochronology to reveal cryptic population histories: review and synthesis of 29 ancient DNA studies. Molecular Ecology, 18(7), 1310-1330.
- LMU BioDB 2019. (2019). Week 5. Retrieved September 30, 2019, from https://xmlpipedb.cs.lmu.edu/biodb/fall2019/index.php/Week_5
- MediaWiki (2019). Category: Help. Retrieved September 30, 2019, from https://www.mediawiki.org/wiki/Category:Help